Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases
Abstract
1. Introduction
2. Results and Discussion
2.1. The Rationale
2.2. Conformational Changes of AP Endonucleases in the Course of F/N-Substrate Cleavage
2.3. Direct PAGE Analysis of F/N-Substrate Cleavage
2.4. Fluorescent Detection of Opposite-Nucleotide Extrusion in the Course of Catalytic-Complex Formation
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. Oligodeoxynucleotides (ODNs)
3.3. Enzyme Assays by Polyacrylamide Gel Electrophoresis (PAGE)
3.4. Stopped-Flow Fluorescence Measurements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dodson, M.L.; Michaels, M.L.; Lloyd, R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994, 269, 32709–32712. [Google Scholar] [CrossRef]
- Dodson, M.L.; Lloyd, R.S. Mechanistic comparison among base exision repair glycosylases. Free Radic. Biol. Med. 2002, 32, 678–682. [Google Scholar] [CrossRef]
- Gros, L.; Saparbaev, M.K.; Laval, J. Enzymology of the repair of free radicals-induced DNA damage. Oncogene 2002, 21, 8905–8925. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schar, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef]
- Golan, G.; Ishchenko, A.A.; Khassenov, B.; Shoham, G.; Saparbaev, M.K. Coupling of the nucleotide incision and 3′→5′ exonuclease activities in Escherichia coli endonuclease IV: Structural and genetic evidences. Mutat. Res. 2010, 685, 70–79. [Google Scholar] [CrossRef]
- Prorok, P.; Saint-Pierre, C.; Gasparutto, D.; Fedorova, O.S.; Ishchenko, A.A.; Leh, H.; Buckle, M.; Tudek, B.; Saparbaev, M. Highly mutagenic exocyclic DNA adducts are substrates for the human nucleotide incision repair pathway. PLoS ONE 2012, 7, e51776. [Google Scholar] [CrossRef]
- Christov, P.P.; Banerjee, S.; Stone, M.P.; Rizzo, C.J. Selective Incision of the alpha-N-Methyl-Formamidopyrimidine Anomer by Escherichia coli Endonuclease IV. J. Nucleic Acids 2010, 2010, 850234. [Google Scholar] [CrossRef]
- Vrouwe, M.G.; Pines, A.; Overmeer, R.M.; Hanada, K.; Mullenders, L.H. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 2011, 124, 435–446. [Google Scholar] [CrossRef]
- Guliaev, A.B.; Hang, B.; Singer, B. Structural insights by molecular dynamics simulations into specificity of the major human AP endonuclease toward the benzene-derived DNA adduct, pBQ-C. Nucleic Acids Res. 2004, 32, 2844–2852. [Google Scholar] [CrossRef]
- Daviet, S.; Couve-Privat, S.; Gros, L.; Shinozuka, K.; Ide, H.; Saparbaev, M.; Ishchenko, A.A. Major oxidative products of cytosine are substrates for the nucleotide incision repair pathway. DNA Repair 2007, 6, 8–18. [Google Scholar] [CrossRef]
- Prorok, P.; Alili, D.; Saint-Pierre, C.; Gasparutto, D.; Zharkov, D.O.; Ishchenko, A.A.; Tudek, B.; Saparbaev, M.K. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2013, 110, E3695–E3703. [Google Scholar] [CrossRef] [PubMed]
- Ischenko, A.A.; Saparbaev, M.K. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 2002, 415, 183–187. [Google Scholar] [CrossRef]
- Ishchenko, A.A.; Sanz, G.; Privezentzev, C.V.; Maksimenko, A.V.; Saparbaev, M. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 2003, 31, 6344–6353. [Google Scholar] [CrossRef]
- Gros, L.; Ishchenko, A.A.; Ide, H.; Elder, R.H.; Saparbaev, M.K. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004, 32, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Barzilay, G.; Walker, L.J.; Robson, C.N.; Hickson, I.D. Site-directed mutagenesis of the human DNA repair enzyme HAP1: Identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 1995, 23, 1544–1550. [Google Scholar] [CrossRef]
- Berquist, B.R.; McNeill, D.R.; Wilson III, D.M. Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J. Mol. Biol 2008, 379, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Barnes, T.; Kim, W.C.; Mantha, A.K.; Kim, S.E.; Izumi, T.; Mitra, S.; Lee, C.H. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009, 37, 3946–3958. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Herman, T.; Demple, B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991, 19, 5907–5914. [Google Scholar] [CrossRef]
- Chou, K.M.; Cheng, Y.C. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3 ’ mispaired DNA. Nature 2002, 415, 655–659. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.A.; Fedorova, O.O.S.O.; Kuznetsov, N.N.A.N. Kinetic Features of 3′–5′ Exonuclease Activity of Human AP-Endonuclease APE1. Molecules 2018, 23, 2101. [Google Scholar] [CrossRef]
- Kerins, S.M.; Collins, R.; McCarthy, T.V. Characterization of an endonuclease IV 3′–5′ exonuclease activity. J. Biol. Chem. 2003, 278, 3048–3054. [Google Scholar] [CrossRef]
- Wilson III, D.M.; Barsky, D. The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001, 485, 283–307. [Google Scholar] [CrossRef]
- Mol, C.D.; Kuo, C.F.; Thayer, M.M.; Cunningham, R.P.; Tainer, J.A. Structure and Function of the Multifunctional DNA-Repair Enzyme Exonuclease-III. Nature 1995, 374, 381–386. [Google Scholar] [CrossRef]
- Hosfield, D.J.; Guan, Y.; Haas, B.J.; Cunningham, R.P.; Tainer, J.A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 1999, 98, 397–408. [Google Scholar] [CrossRef]
- Mol, C.D.; Hosfield, D.J.; Tainer, J.A. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: The 3′ ends justify the means. Mutat. Res. 2000, 460, 211–229. [Google Scholar] [CrossRef]
- Garcin, E.D.; Hosfield, D.J.; Desai, S.A.; Haas, B.J.; Bjoras, M.; Cunningham, R.P.; Tainer, J.A. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat. Struct. Mol. Biol. 2008, 15, 515–522. [Google Scholar] [CrossRef]
- Mol, C.D.; Izumi, T.; Mitra, S.; Tainer, J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 2000, 403, 451–456. [Google Scholar] [CrossRef]
- Tsutakawa, S.E.; Shin, D.S.; Mol, C.D.; Izumi, T.; Arvai, A.S.; Mantha, A.K.; Szczesny, B.; Ivanov, I.N.; Hosfield, D.J.; Maiti, B.; et al. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem. 2013, 288, 8445–8455. [Google Scholar] [CrossRef]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Vorobjev, Y.N.; Kuznetsov, N.A.; Fedorova, O.S. Effects of mono- and divalent metal ions on DNA binding and catalysis of human apurinic/apyrimidinic endonuclease 1. Mol. BioSyst. 2016, 12, 1527–1539. [Google Scholar] [CrossRef]
- Kanazhevskaya, L.Y.; Koval, V.V.; Zharkov, D.O.; Strauss, P.R.; Fedorova, O.S. Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair. Biochemistry 2010, 49, 6451–6461. [Google Scholar] [CrossRef]
- Timofeyeva, N.A.; Koval, V.V.; Knorre, D.G.; Zharkov, D.O.; Saparbaev, M.K.; Ishchenko, A.A.; Fedorova, O.S. Conformational dynamics of human AP endonuclease in base excision and nucleotide incision repair pathways. J. Biomol. Struct. Dyn. 2009, 26, 637–652. [Google Scholar] [CrossRef]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Kuznetsov, N.A.; Fedorova, O.S. Thermodynamics of Damaged DNA Binding and Catalysis by Human AP Endonuclease 1. Acta Nat. 2016, 8, 103–110. [Google Scholar] [CrossRef]
- Zang, H.; Fang, Q.; Pegg, A.E.; Guengerich, F.P. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2005, 280, 30873–30881. [Google Scholar] [CrossRef]
- Yang, K.; Stanley, R.J. The extent of DNA deformation in DNA photolyase-substrate complexes: A solution state fluorescence study. Photochem. Photobiol. 2008, 84, 741–749. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Vorobjev, Y.N.; Krasnoperov, L.N.; Fedorova, O.S. Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence—stopped-flow pre-steady-state kinetics. Nucleic Acids Res. 2012, 40, 7384–7392. [Google Scholar] [CrossRef]
- Kladova, O.A.; Kuznetsov, N.A.; Fedorova, O.S. Thermodynamics of the DNA repair process by endonuclease VIII. Acta Nat. 2019, 11, 29–37. [Google Scholar] [CrossRef]
- Kladova, O.A.; Krasnoperov, L.N.; Kuznetsov, N.A.; Fedorova, O.S. Kinetics and thermodynamics of DNA processing by wild type DNA-glycosylase endo III and its catalytically inactive mutant forms. Genes 2018, 9, 190. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Vorobjev, Y.N.; Barthes, N.P.F.; Michel, B.Y.; Burger, A.; Fedorova, O.S. New Environment-Sensitive Multichannel DNA Fluorescent Label for Investigation of the Protein-DNA Interactions. PLoS ONE 2014, 9, e100007. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kladova, O.A.; Barthes, N.P.F.; Michel, B.Y.; Burger, A.; Fedorova, O.S.; Kuznetsov, N.A. Comparative Analysis of Nucleotide Fluorescent Analogs for Registration of DNA Conformational Changes Induced by Interaction with Formamidopyrimidine-DNA Glycosylase Fpg. Russ. J. Bioorgan. Chem. 2019, 45. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kuznetsov, N.A.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Pre-steady-state fluorescence analysis of damaged DNA transfer from human DNA glycosylases to AP endonuclease APE1. Biochim. Biophys. Acta 2014, 1840, 3042–3051. [Google Scholar] [CrossRef]
- Harlow, E.; Lane, D. Bradford Assay. Cold Spring Harb. Protoc. 2006, 2006, 4644. [Google Scholar] [CrossRef]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Koval, V.V.; Nevinsky, G.A.; Douglas, K.T.; Zharkov, D.O.; Fedorova, O.S. Kinetic conformational analysis of human 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2007, 282, 1029–1038. [Google Scholar] [CrossRef]
- Alekseeva, I.V.; Bakman, A.S.; Vorobjev, Y.N.; Fedorova, O.S.; Kuznetsov, N.A. Role of Ionizing Amino Acid Residues in the Process of DNA Binding by Human AP Endonuclease 1 and in Its Catalysis. J. Phys. Chem. 2019, 123, 9546–9556. [Google Scholar] [CrossRef]
- Yakovlev, D.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Search for Modified DNA Sites with the Human Methyl-CpG-Binding Enzyme MBD4. Acta Nat. 2017, 9, 88–98. [Google Scholar] [CrossRef]
- Alekseeva, I.V.; Kuznetsova, A.A.; Bakman, A.S.; Fedorova, O.S.; Kuznetsov, N.A. The role of active-site amino acid residues in the cleavage of DNA and RNA substrates by human apurinic/apyrimidinic endonuclease APE1. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129718. [Google Scholar] [CrossRef]
- Kladova, O.A.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Mutational and Kinetic Analysis of Lesion Recognition by Escherichia coli Endonuclease VIII. Genes 2017, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Fedorova, O.S. Kinetic milestones of damage recognition by DNA glycosylases of the Helix-hairpin-Helix structural superfamily. Adv. Exp. Biol. Med. 2020, 1241, 1–18. [Google Scholar]
- Kuznetsova, A.A.; Matveeva, A.G.; Milov, A.D.; Vorobjev, Y.N.; Dzuba, S.A.; Fedorova, O.S.; Kuznetsov, N.A. Substrate specificity of human apurinic/apyrimidinic endonuclease APE1 in the nucleotide incision repair pathway. Nucleic Acids Res. 2018, 46, 11454–11465. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Functions of disordered regions in mammalian early base excision repair proteins. Cell. Mol. Life Sci. 2010, 67, 3573. [Google Scholar] [CrossRef]
- Antoniali, G.; Lirussi, L.; Poletto, M.; Tell, G. Emerging roles of the nucleolus in regulating the DNA damage response: The noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example. Antioxid. Redox Signal. 2014, 20, 621–639. [Google Scholar] [CrossRef]
- Davletgildeeva, A.T.; Ishchenko, A.A.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. The Enigma of Substrate Recognition and Catalytic Efficiency of APE1-Like Enzymes. Front. Cell Dev. Biol. 2021, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Fantini, D.; Vascotto, C.; Marasco, D.; D’Ambrosio, C.; Romanello, M.; Vitagliano, L.; Pedone, C.; Poletto, M.; Cesaratto, L.; Quadrifoglio, F.; et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res. 2010, 38, 8239–8256. [Google Scholar] [CrossRef]
- Poletto, M.; Vascotto, C.; Scognamiglio, P.L.; Lirussi, L.; Marascod, D.; Tell, G. Role of the unstructured N-terminal domain of the hAPE1 (human apurinic/apyrimidinic endonuclease 1) in the modulation of its interaction with nucleic acids and NPM1 (nucleophosmin). Biochem. J. 2013, 452, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, R.; Wiederhold, L.; Szczesny, B.; Boldogh, I.; Hazra, T.K.; Izumi, T.; Mitra, S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006, 34, 2067–2076. [Google Scholar] [CrossRef]
- Izumi, T.; Mitra, S. Deletion analysis of human AP-endonuclease: Minimum sequence required for the endonuclease activity. Carcinogenesis 1998, 19, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Kladova, O.A.; Bazlekowa-Karaban, M.; Baconnais, S.; Piétrement, O.; Ishchenko, A.A.; Matkarimov, B.T.; Iakovlev, D.A.; Vasenko, A.; Fedorova, O.S.; Le Cam, E.; et al. The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation. DNA Repair 2018, 64, 10–25. [Google Scholar] [CrossRef]
- Moor, N.; Vasil’eva, I.; Lavrik, O. Functional role of N-terminal extension of human ap endonuclease 1 in coordination of base excision dna repair via protein–protein interactions. Int. J. Mol. Sci. 2020, 21, 3122. [Google Scholar] [CrossRef]
- Popov, A.V.; Grin, I.R.; Dvornikova, A.P.; Matkarimov, B.T.; Groisman, R.; Saparbaev, M.; Zharkov, D.O. Reading Targeted DNA Damage in the Active Demethylation Pathway: Role of Accessory Domains of Eukaryotic AP Endonucleases and Thymine-DNA Glycosylases. J. Mol. Biol. 2020, 432, 1747–1768. [Google Scholar] [CrossRef]
- López, D.J.; Rodríguez, J.A.; Bañuelos, S. Molecular Mechanisms Regulating the DNA Repair Protein APE1: A Focus on Its Flexible N-Terminal Tail Domain. Int. J. Mol. Sci. 2021, 22, 6308. [Google Scholar] [CrossRef] [PubMed]
- Bazlekowa-Karaban, M.; Prorok, P.; Baconnais, S.; Taipakova, S.; Akishev, Z.; Zembrzuska, D.; Popov, A.V.; Endutkin, A.V.; Groisman, R.; Ishchenko, A.A.; et al. Mechanism of stimulation of DNA binding of the transcription factors by human apurinic/apyrimidinic endonuclease 1, APE1. DNA Repair 2019, 82, 102698. [Google Scholar] [CrossRef]
- Kelley, M.R.; Georgiadis, M.M.; Fishel, M.L. APE1/Ref-1Role in Redox Signaling: Translational Applications of Targeting the Redox Function of the DNA Repair/Redox Protein APE1/Ref-1. Curr. Mol. Pharmacol. 2011, 5, 36–53. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, M.M.; Luo, M.; Gaur, R.K.; Delaplane, S.; Li, X.; Kelley, M.R. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2008, 643, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Vascotto, C.; Fantini, D.; Romanello, M.; Cesaratto, L.; Deganuto, M.; Leonardi, A.; Radicella, J.P.; Kelley, M.R.; D’Ambrosio, C.; Scaloni, A.; et al. APE1/Ref-1 Interacts with NPM1 within Nucleoli and Plays a Role in the rRNA Quality Control Process. Mol. Cell. Biol. 2009, 29, 1834–1854. [Google Scholar] [CrossRef]
- Tell, G.; Wilson, D.M.; Lee, C.H. Intrusion of a DNA Repair Protein in the RNome World: Is This the Beginning of a New Era? Mol. Cell. Biol. 2010, 30, 366–371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antoniali, G.; Serra, F.; Lirussi, L.; Tanaka, M.; D’Ambrosio, C.; Zhang, S.; Radovic, S.; Dalla, E.; Ciani, Y.; Scaloni, A.; et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat. Commun. 2017, 8, 797. [Google Scholar] [CrossRef]
- Takeuchi, M.; Lillis, R.; Demple, B.; Takeshita, M. Interactions of Escherichia coli Endonuclease IV and Exonuclease III with Abasic Sites in DNA. J. Biol. Chem. 1994, 269, 21907–21914. [Google Scholar] [CrossRef]



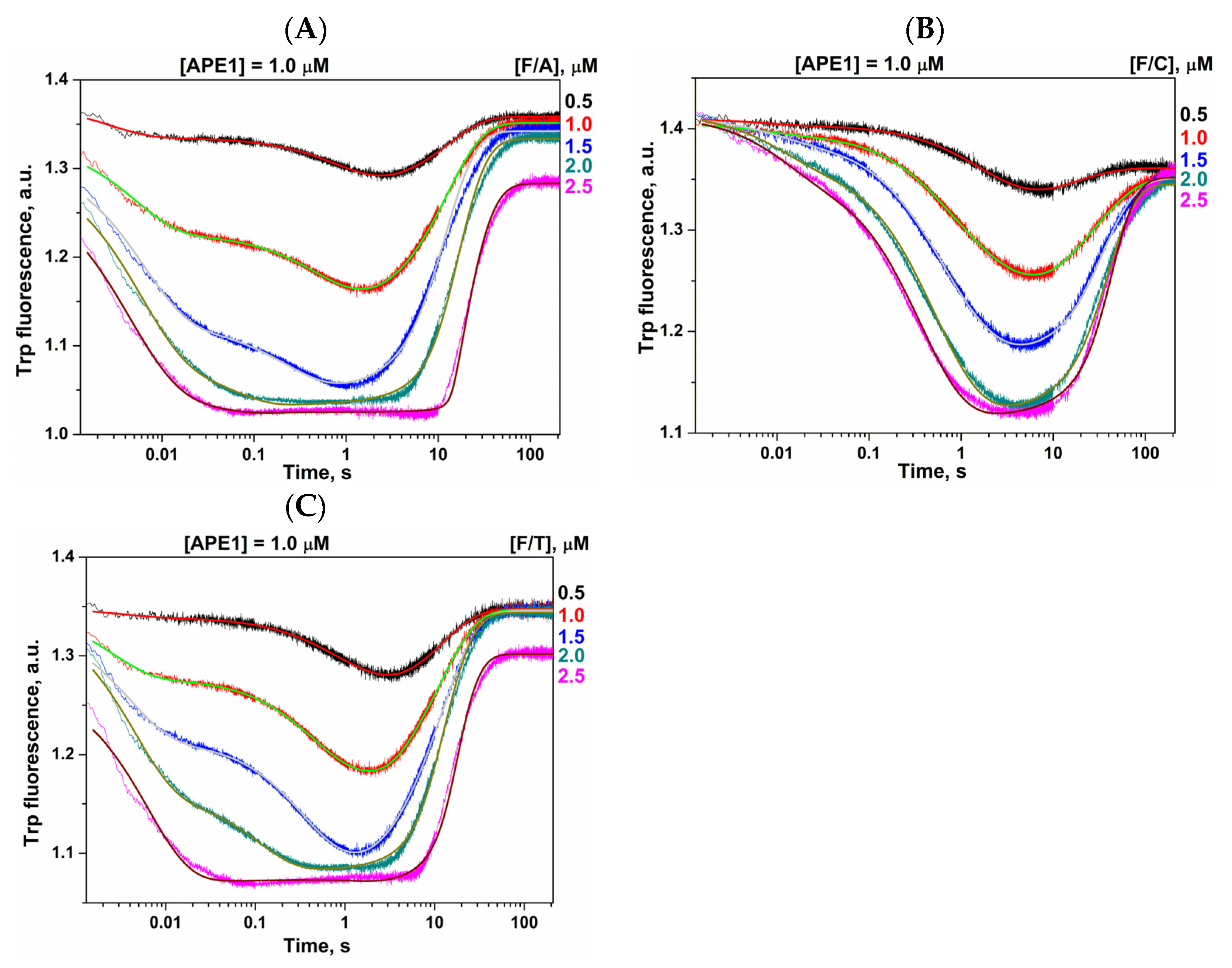

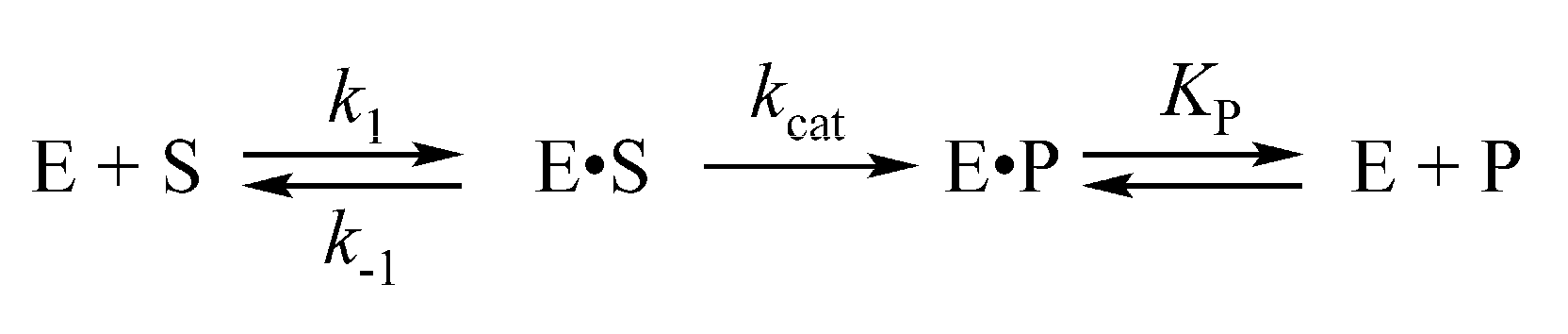

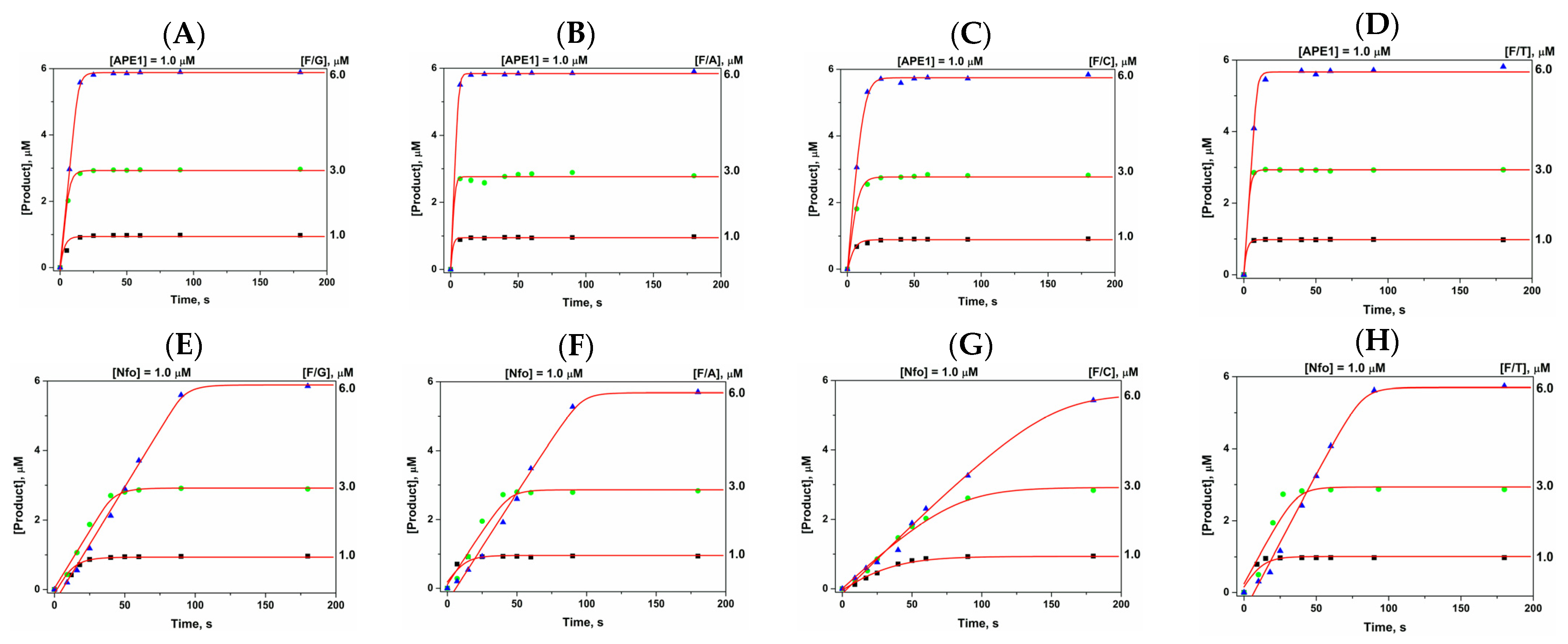
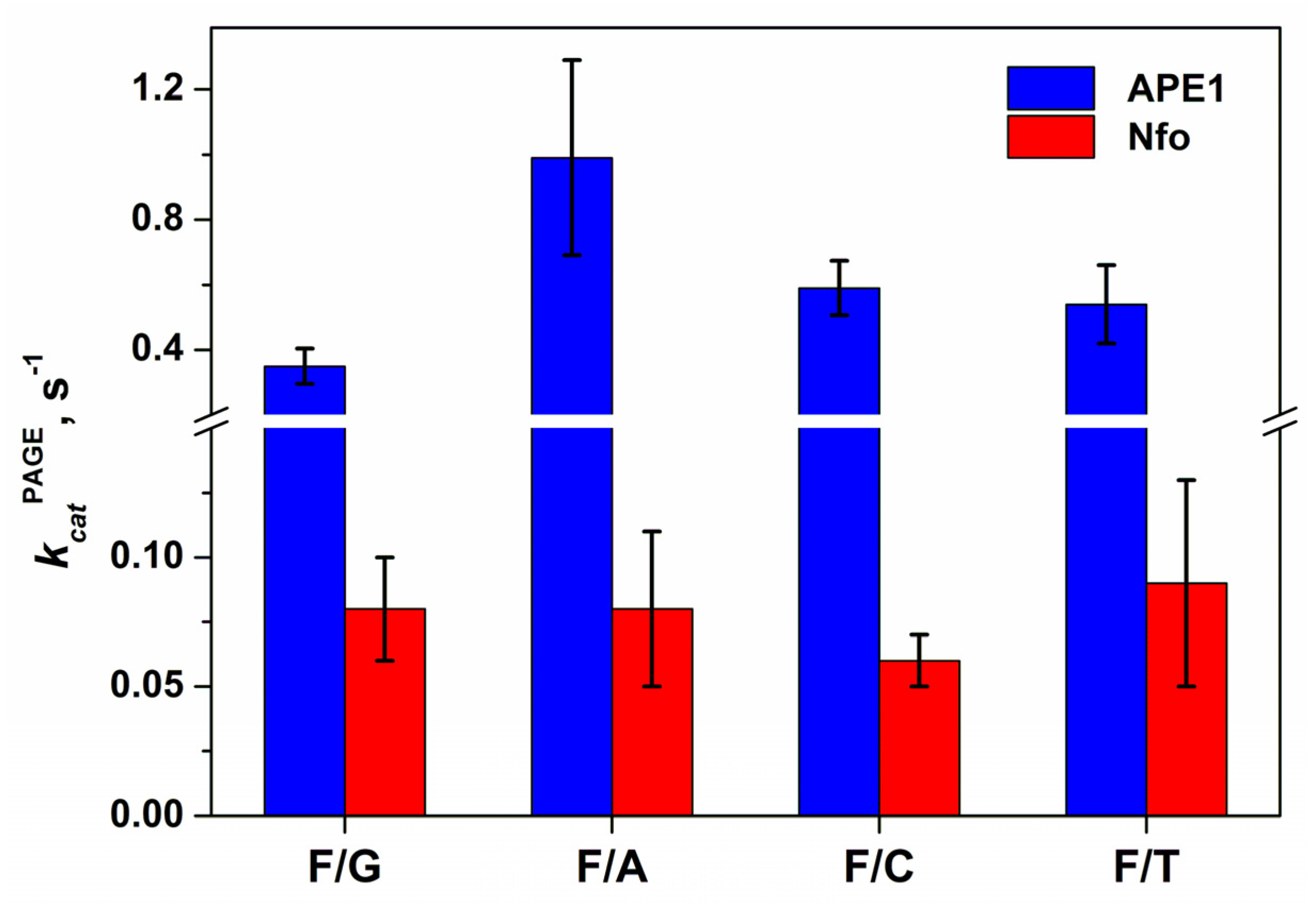

| Constants | F/G b | F/A | F/C | F/T |
|---|---|---|---|---|
| k1, M−1s−1 | (90 ± 10) × 106 | (37 ± 4) × 106 | (5.5 ± 1.2) × 106 | (58 ± 7) × 106 |
| k−1, s−1 | 10.1 ± 4.3 | 66 ± 9 | 110 ± 10 | 120 ± 10 |
| K1, M−1 | (8.9 ± 4.8) × 106 | (0.6 ± 0.1) × 106 | (0.05 ± 0.01) × 106 | (0.5 ± 0.1) × 106 |
| k2, s−1 | 43.1 ± 8.5 | 3.9 ± 0.3 | 11.5 ± 2.7 | 10.0 ± 2.0 |
| k−2, s−1 | 7.1 ± 1.6 | 0.47 ± 0.02 | 0.52 ± 0.02 | 0.88 ± 0.06 |
| K2 | 6.1 ± 2.6 | 8.3 ± 1.0 | 22 ± 6 | 11 ± 3 |
| Kbind, M−1 | (63 ± 57) × 106 | (5.6 ± 1.5) × 106 | (1.2 ± 0.5) × 106 | (6.0 ± 2.7) × 106 |
| kcat, s−1 | 0.3 ± 0.1 | 0.16 ± 0.01 | 0.10 ± 0.01 | 0.23 ± 0.03 |
| KP, M | (7.2 ± 0.4) × 10−6 | (1.0 ± 0.2) × 10−6 | (11 ± 2) × 10−6 | (32 ± 5) × 10−6 |
| Constants | F/G | F/A | F/C | F/T |
|---|---|---|---|---|
| k1, M−1s−1 | (151 ± 2) × 106 | (360 ± 10) × 106 | (150 ± 10) × 106 | (360 ± 10) × 106 |
| k−1, s−1 | 1.9 ± 0.2 | 30 ± 1 | 270 ± 10 | 235 ± 10 |
| K1, M−1 | (80 ± 9) × 106 | (12 ± 1) × 106 | (0.55 ± 0.06) × 106 | (1.5 ± 0.1) × 106 |
| k2, s−1 | - | - | 9.1 ± 0.2 | 101 ± 2 |
| k−2, s−1 | - | - | 4.4 ± 0.1 | 28 ± 1 |
| K2 | - | - | 2.1 ± 0.1 | 3.6 ± 0.2 |
| Kbind, M−1 | (80 ± 9) × 106 | (12 ± 1) × 106 | (1.7 ± 0.2) × 106 | (6.9 ± 0.8) × 106 |
| kcat, s−1 | 0.10 ± 0.01 | 0.18 ± 0.02 | 0.27 ± 0.01 | 0.23 ± 0.01 |
| KP, M | (11 ± 1) × 10−6 | (5.8 ± 0.4) × 10−6 | (6.9 ± 0.4) × 10−6 | (2.7 ± 0.1) × 10−6 |
| Shorthand | Sequences of DNA Duplexes |
|---|---|
| F/N-substrate N = G, A, T, C | 5’-CTCTCFCCTTCC-3’ 3’-GAGAGNGGAAGG-5’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, A.A.; Senchurova, S.I.; Ishchenko, A.A.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases. Int. J. Mol. Sci. 2021, 22, 8874. https://doi.org/10.3390/ijms22168874
Kuznetsova AA, Senchurova SI, Ishchenko AA, Saparbaev M, Fedorova OS, Kuznetsov NA. Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases. International Journal of Molecular Sciences. 2021; 22(16):8874. https://doi.org/10.3390/ijms22168874
Chicago/Turabian StyleKuznetsova, Alexandra A., Svetlana I. Senchurova, Alexander A. Ishchenko, Murat Saparbaev, Olga S. Fedorova, and Nikita A. Kuznetsov. 2021. "Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases" International Journal of Molecular Sciences 22, no. 16: 8874. https://doi.org/10.3390/ijms22168874
APA StyleKuznetsova, A. A., Senchurova, S. I., Ishchenko, A. A., Saparbaev, M., Fedorova, O. S., & Kuznetsov, N. A. (2021). Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases. International Journal of Molecular Sciences, 22(16), 8874. https://doi.org/10.3390/ijms22168874







