Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics
Abstract
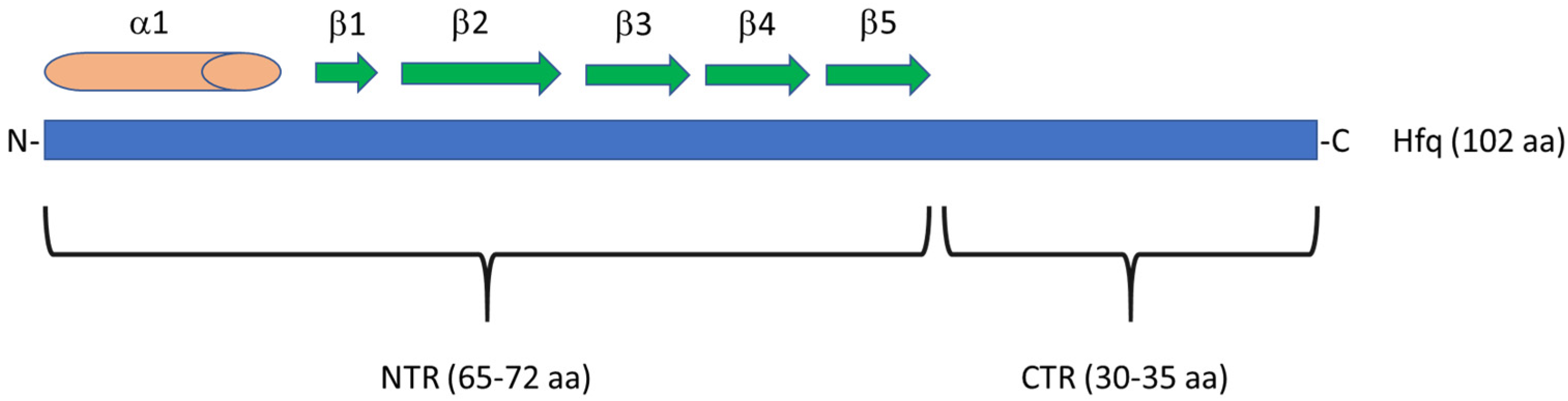
1. Introduction
2. Results
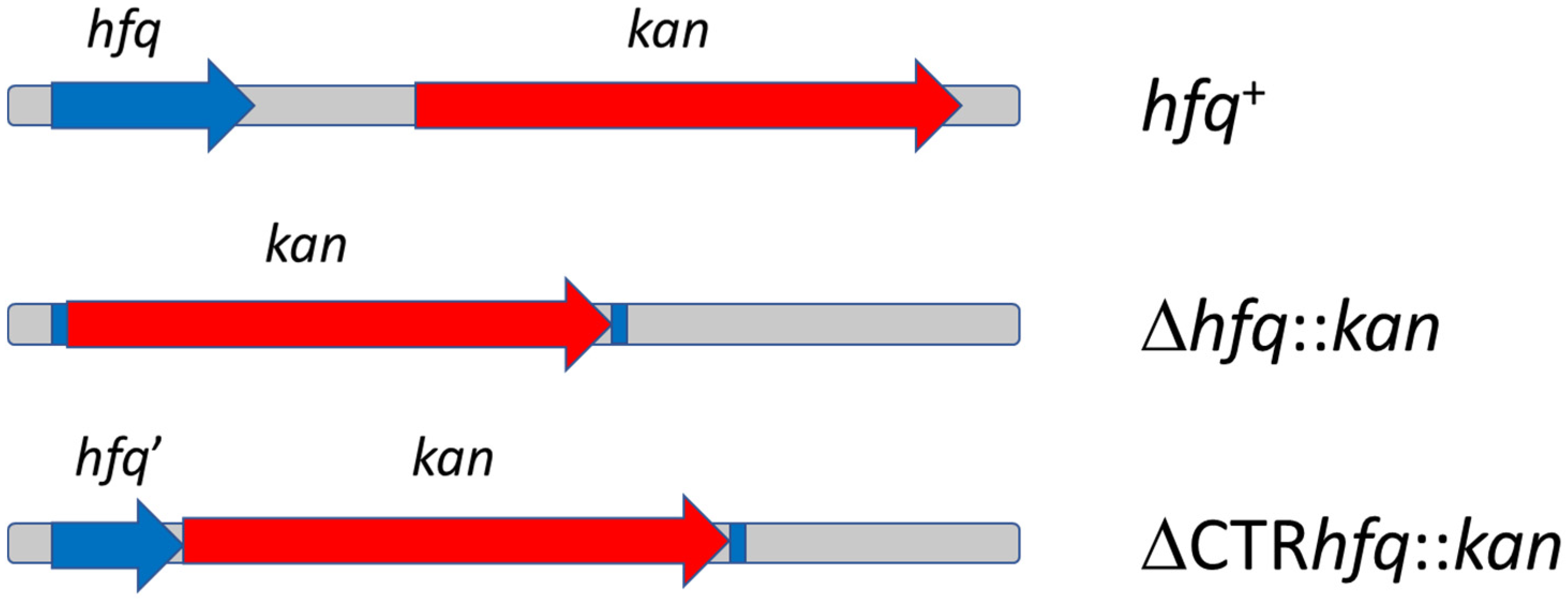
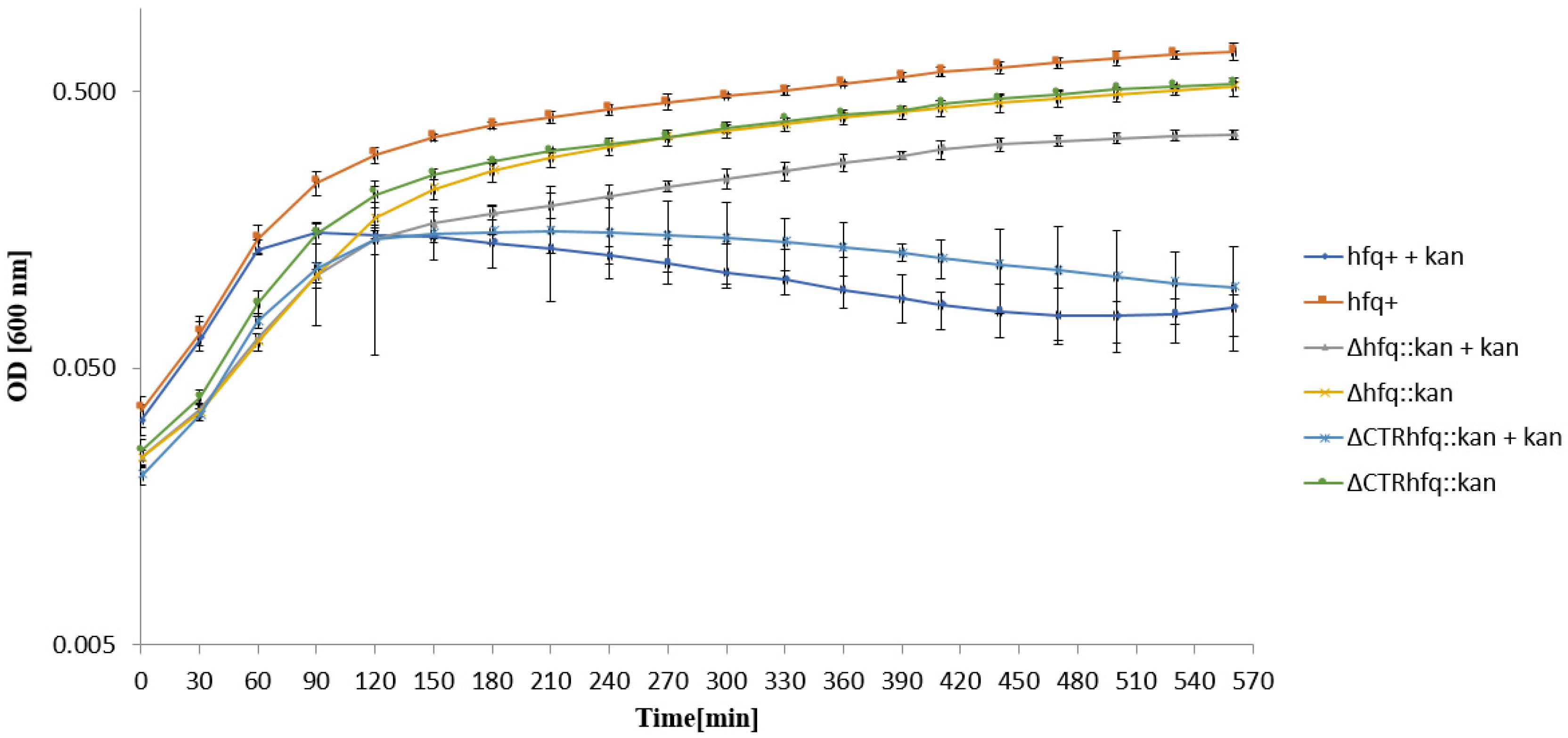
2.1. Chromosome-Borne Resistance to Kanamycin
2.2. Transformation Efficiency of hfq+ and hfq Mutant Hosts with Plasmids
2.3. Low Copy Number Plasmid p27cmr (Lambdoid)-Borne Resistance to Chloramphenicol
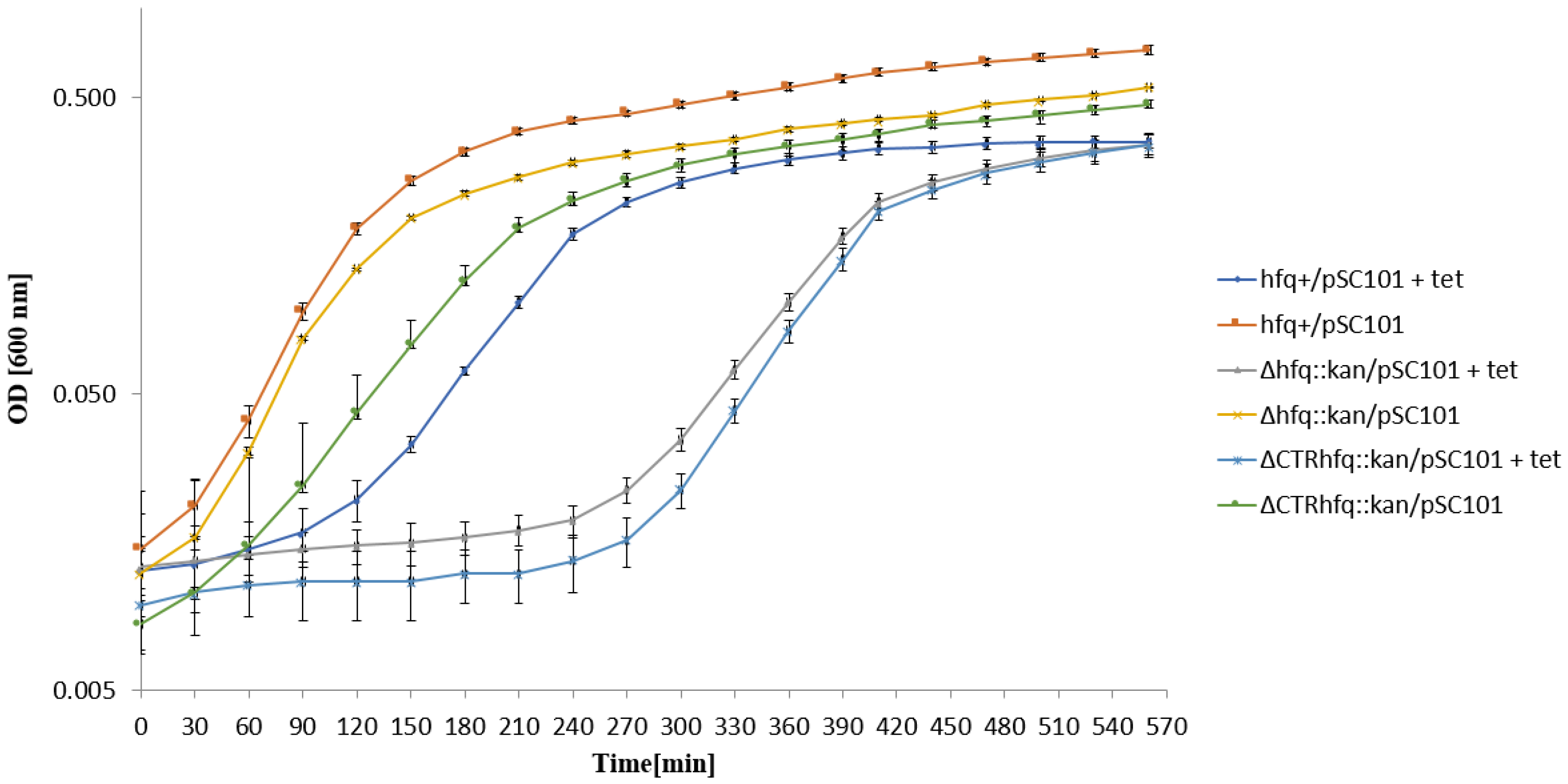
2.4. Low Copy Number Plasmid pSC101-Borne Resistance to Tetracycline
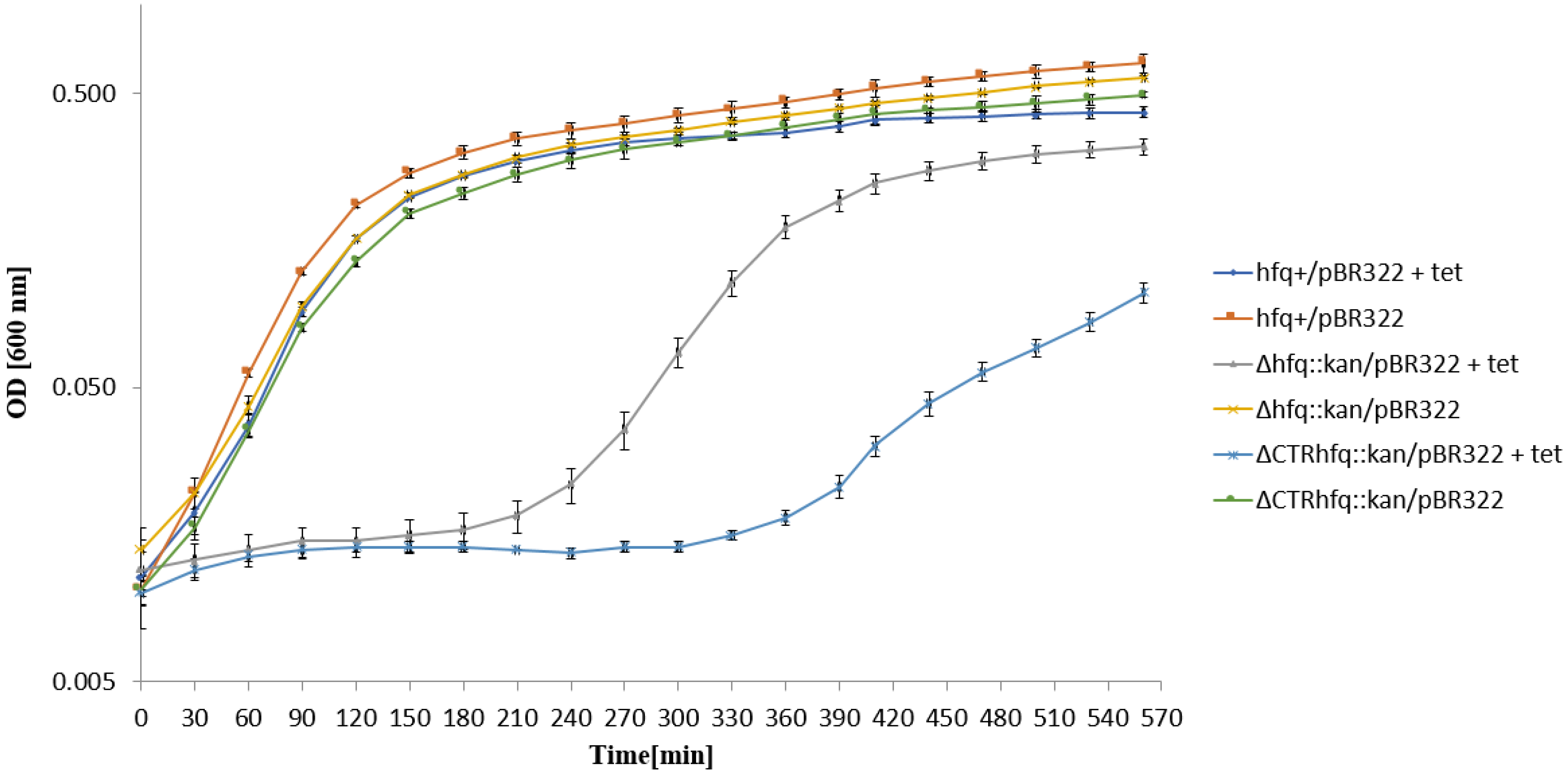
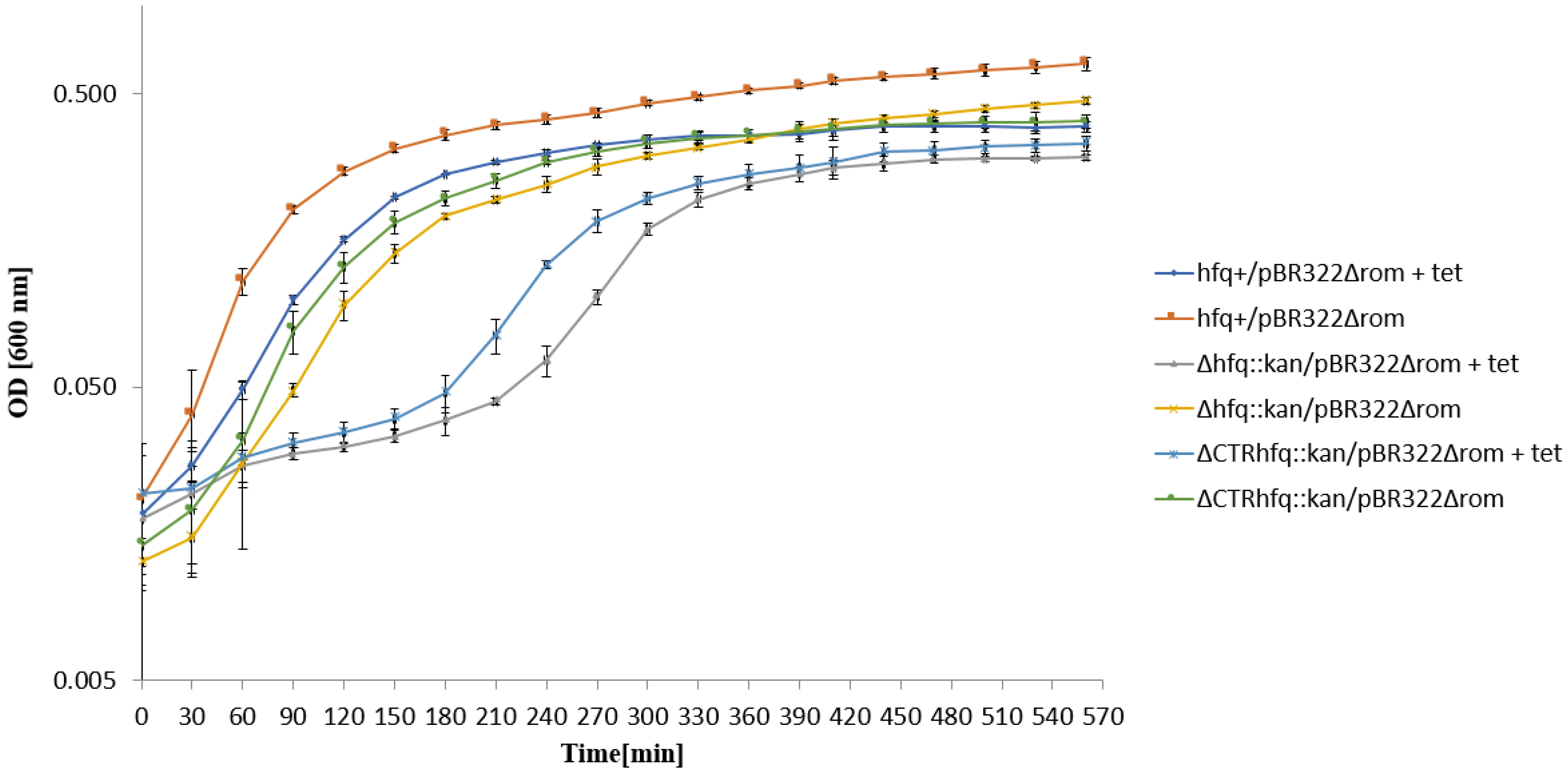
2.5. Medium and High Copy Number ColE1-like Plasmid-Borne Resistance to Tetracycline
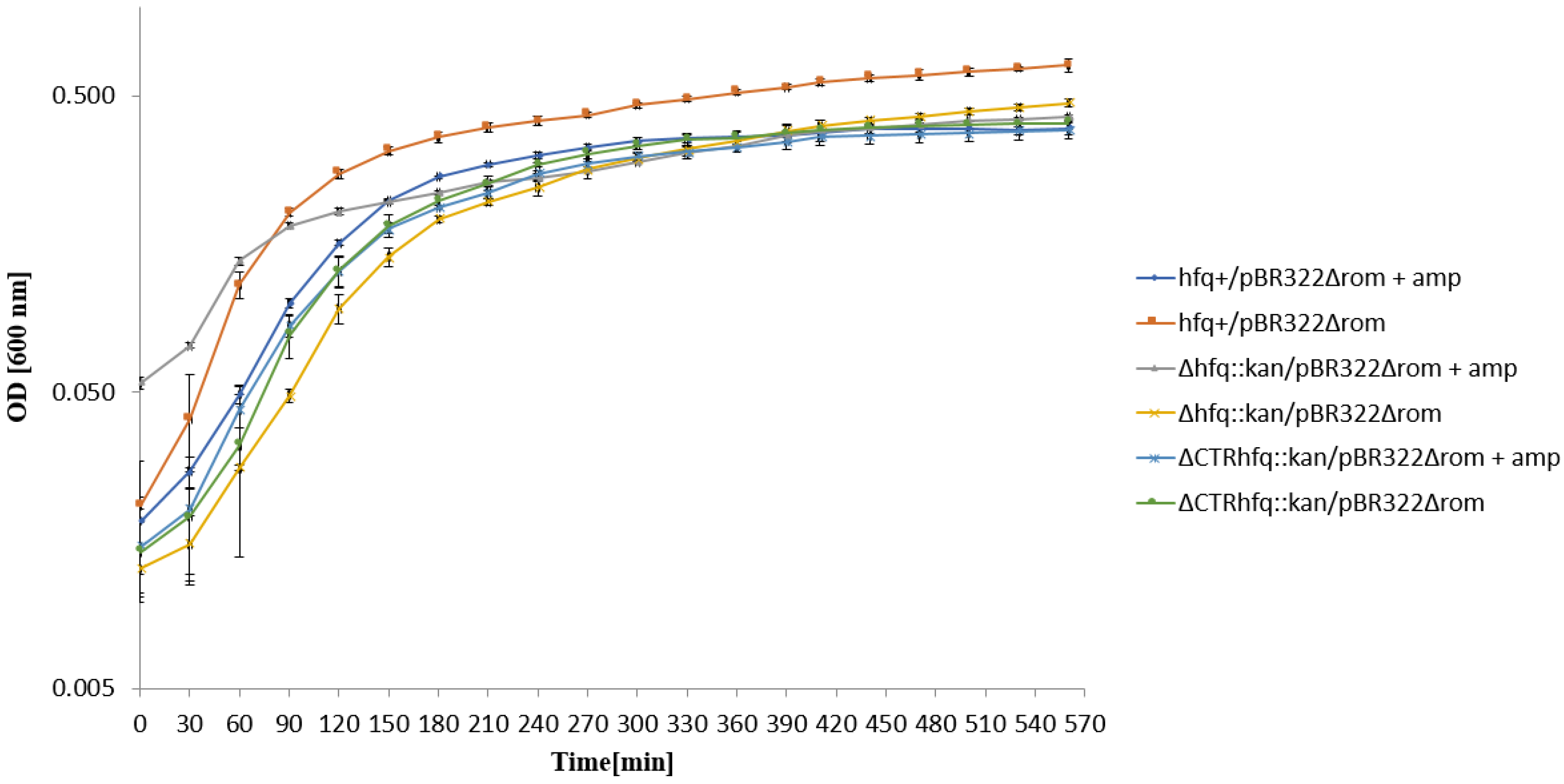
2.6. Medium and High Copy Number ColE1-like Plasmid-Borne Resistance to Ampicillin
2.7. Copy Number of Plasmid pBR322 in hfq+ and hfq Mutant Hosts at Various Growth Phases
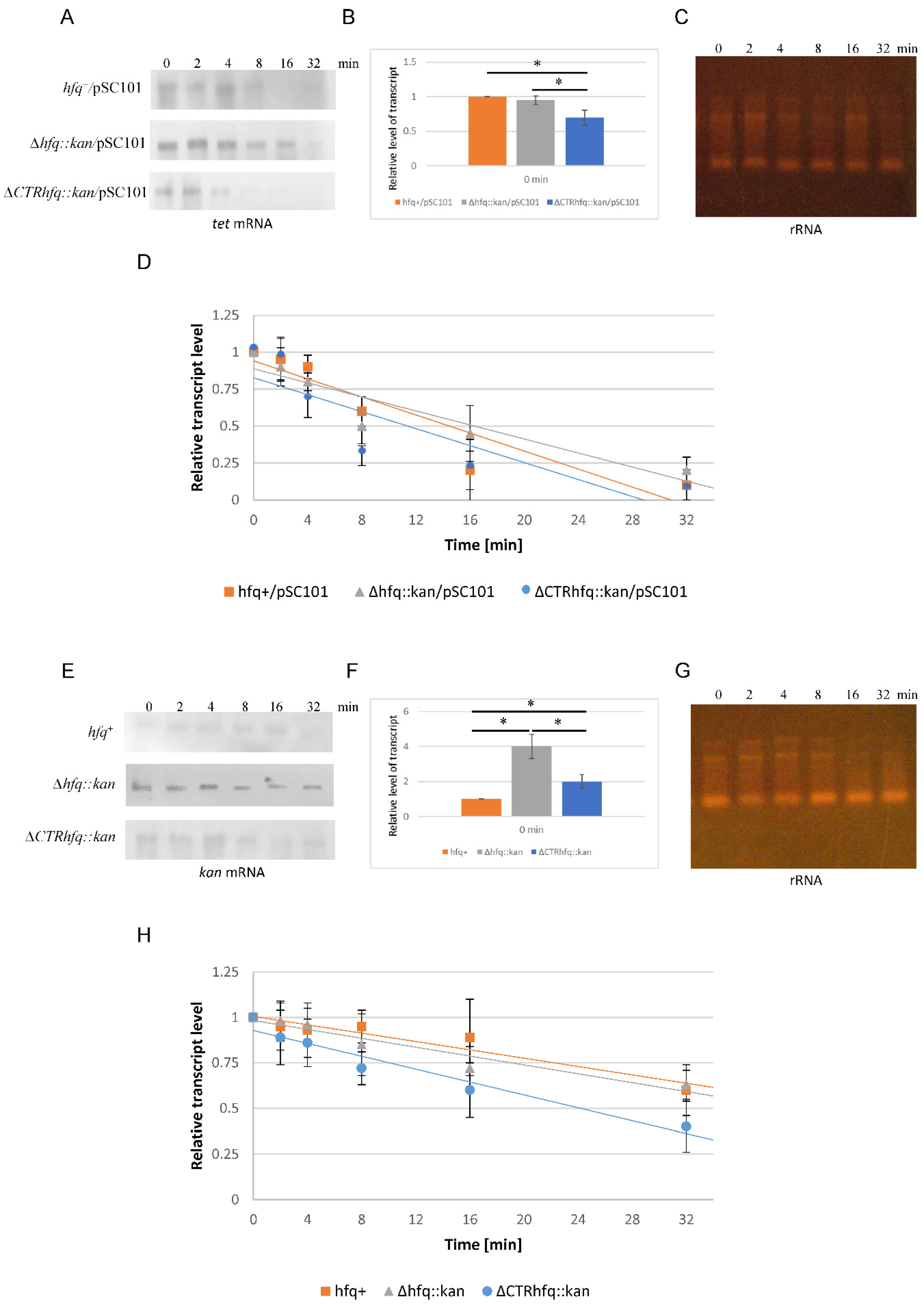
2.8. Decay of mRNAs Derived from Tet and Kan Genes in hfq+ and hfq Mutant Hosts
2.9. Levels of Hfq and NTRHfq Proetins in hfq+ and ΔCTRhfq::kan Strains, Respectively
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Plasmids
4.3. Media and Growth Conditions
4.4. Antibiotic Resistance Estimated on Plates with Solid Medium
4.5. Monitoring of Growth in Liquid Medium
4.6. Efficiency of Transformation
4.7. Plasmid Copy Number
4.8. Northern-Blotting
4.9. Determination of Minimal Inhibitory Concentration (MIC) of Antibiotics
4.10. Determination of Levels of Hfq Proteins
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franze De Fernandez, M.T.; Hayward, W.S.; August, J.T. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972, 247, 824–831. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. RNA reflections: Converging on Hfq. RNA 2015, 21, 511–512. [Google Scholar] [CrossRef]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef]
- Dos Santos, R.F.; Arraiano, C.M.; Andrade, J.M. New molecular interactions broaden the functions of the RNA chaperone Hfq. Curr. Genet. 2019, 65, 1313–1319. [Google Scholar]
- Cech, G.M.; Szalewska-Pałasz, A.; Kubiak, K.; Malabirade, A.; Grange, W.; Arluison, V.; Węgrzyn, G. The Escherichia coli Hfq protein: An unattended DNA-transactions regulator. Front. Mol. Biosci. 2016, 3, 36. [Google Scholar] [CrossRef]
- Santiago-Frangos, A.; Kavita, K.; Schu, D.; Gottesman, S.; Woodson, S.A. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. USA 2016, 113, E6089–E6096. [Google Scholar] [CrossRef]
- Santiago-Frangos, A.; Fröhlich, K.S.; Jeliazkov, J.R.; Małecka, E.M.; Marino, G.; Gray, J.J.; Luisi, B.F.; Woodson, S.A.; Hardwick, S. Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation. Proc. Natl. Acad. Sci. USA 2019, 116, 10978–10987. [Google Scholar] [CrossRef] [PubMed]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trépout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35, e00190. [Google Scholar] [CrossRef]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef] [PubMed]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef] [PubMed]
- Cech, G.M.; Pakuła, B.; Kamrowska, D.; Węgrzyn, G.; Arluison, V.; Szalewska-Pałasz, A. Hfq protein deficiency in Escherichia coli affects ColE1-like but not lambda plasmid DNA replication. Plasmid 2014, 73, 10–15. [Google Scholar] [CrossRef]
- Wien, F.; Martinez, D.; Le Brun, E.; Jones, N.C.; Vrønning Hoffmann, S.; Waeytens, J.; Berbon, M.; Habenstein, B.; Arluison, V. The bacterial amyloid-like Hfq promotes in vitro DNA alignment. Microorganisms 2019, 7, 639. [Google Scholar] [CrossRef]
- Parekh, V.J.; Niccum, B.A.; Shah, R.; Rivera, M.A.; Novak, M.J.; Geinguenaud, F.; Wien, F.; Arluison, V.; Sinden, R.R. Role of Hfq in genome evolution: Instability of G-quadruplex sequences in E. coli. Microorganisms 2020, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- El Hamoui, O.; Yadav, I.; Radiom, M.; Wien, F.; Berret, J.F.; van der Maarel, J.R.C.; Arluison, V. Interactions between DNA and the Hfq amyloid-like region trigger a viscoelastic response. Biomacromolecules 2020, 21, 3668–3677. [Google Scholar] [CrossRef] [PubMed]
- Parekh, V.J.; Wien, F.; Grange, W.; De Long, T.A.; Arluison, V.; Sinden, R.R. Crucial role of the C-terminal domain of Hfq protein in genomic instability. Microorganisms 2020, 8, 1598. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Song, Y.; Yang, L.; Feng, Y.; Qiu, Y.; Li, G.; Guo, J.; Bi, Y.; Qu, Y.; Wang, W.; et al. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE 2009, 4, e6213. [Google Scholar] [CrossRef] [PubMed]
- Correia Santos, S.; Bischler, T.; Westermann, A.J.; Vogel, J. MAPS integrates regulation of actin-targeting effector SteC into the virulence control network of Salmonella small RNA PinT. Cell Rep. 2021, 34, 108722. [Google Scholar] [CrossRef] [PubMed]
- Corsi, I.D.; Dutta, S.; van Hoof, A.; Koehler, T.M. AtxA-controlled small RNAs of Bacillus anthracis virulence plasmid pXO1 regulate gene xxpression in trans. Front. Microbiol. 2021, 11, 610036. [Google Scholar] [CrossRef]
- Crispim, J.S.; da Silva, T.F.; Sanches, N.M.; da Silva, G.C.; Pereira, M.F.; Rossi, C.C.; Li, Y.; Terra, V.S.; Vohra, P.; Wren, B.W.; et al. Serovar-dependent differences in Hfq-regulated phenotypes in Actinobacillus pleuropneumoniae. Pathog. Dis. 2020, 78, ftaa066. [Google Scholar] [CrossRef]
- Dienstbier, A.; Amman, F.; Štipl, D.; Petráčková, D.; Večerek, B. Comparative integrated omics analysis of the Hfq regulon in Bordetella pertussis. Int. J. Mol. Sci. 2019, 20, 3073. [Google Scholar] [CrossRef]
- Alvarez Hayes, J.; Surmann, K.; Lamberti, Y.; Depke, M.; Dhople, V.; Blancá, B.; Ruiz, E.; Vecerek, B.; Schmidt, F.; Völker, U.; et al. Hfq modulates global protein pattern and stress response in Bordetella pertussis. J. Proteomics 2020, 211, 103559. [Google Scholar] [CrossRef] [PubMed]
- Hill, I.T.; Tallo, T.; Dorman, M.J.; Dove, S.L. Loss of RNA chaperone Hfq unveils a toxic pathway in Pseudomonas aeruginosa. J. Bacteriol. 2019, 201, e00232. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Kang, M.; Wang, Y.; Feng, Y.; Kong, S.; Cai, X.; Ling, Z.; Chen, S.; Jiao, X.; Yin, Y. An essential role for hfq involved in biofilm formation and virulence in serotype 4b Listeria monocytogenes. Microbiol. Res. 2018, 215, 148–154. [Google Scholar] [CrossRef]
- Yamada, J.; Yamasaki, S.; Hirakawa, H.; Hayashi-Nishino, M.; Yamaguchi, A.; Nishino, K. Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli. J. Antimicrob. Chemother. 2010, 65, 853–858. [Google Scholar] [CrossRef]
- Kim, T.; Bak, G.; Lee, J.; Kim, K.S. Systematic analysis of the role of bacterial Hfq-interacting sRNAs in the response to antibiotics. J. Antimicrob. Chemother. 2015, 70, 1659–1968. [Google Scholar] [CrossRef] [PubMed]
- Dersch, P.; Khan, M.A.; Mühlen, S.; Görke, B. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 2017, 8, 803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, W.; Tang, Y.; Li, H.; Ma, X.; Liu, Z. RNA chaperone hfq mediates persistence to multiple antibiotics in Aeromonas veronii. Microb. Pathog. 2019, 132, 124–128. [Google Scholar] [CrossRef]
- Pusic, P.; Sonnleitner, E.; Krennmayr, B.; Heitzinger, D.A.; Wolfinger, M.T.; Resch, A.; Bläsi, U. Harnessing metabolic regulation to increase Hfq-dependent antibiotic susceptibility in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 2709. [Google Scholar] [CrossRef]
- Andrade, J.M.; Dos Santos, R.F.; Chelysheva, I.; Ignatova, Z.; Arraiano, C.M. The RNA-binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J. 2018, 37, e97631. [Google Scholar] [CrossRef]
- Turbant, F.; Partouche, D.; El Hamoui, O.; Trépout, S.; Legoubey, T.; Wien, F.; Arluison, V. Apomorphine targets the pleiotropic bacterial regulator Hfq. Antibiotics 2021, 10, 257. [Google Scholar] [CrossRef]
- Brauner, A.; Friedman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Brantl, S. Plasmid replication control by antisense RNAs. Microbiol. Spectr. 2014, 2, PLAS-0001-2013. [Google Scholar] [CrossRef]
- Lilly, J.; Camps, M. Mechanisms of theta plasmid replication. Microbio. Spectr. 2015, 3, PLAS-0029-2014. [Google Scholar] [CrossRef]
- Ross, J.A.; Trussler, R.S.; Black, M.D.; McLellan, C.R.; Haniford, D.B. Tn5 transposition in Escherichia coli is repressed by Hfq and activated by over-expression of the small non-coding RNA SgrS. Mob. DNA 2014, 5, 27. [Google Scholar] [CrossRef]
- Ellis, M.J.; Trussler, R.S.; Haniford, D.B. Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 2015, 96, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Le Derout, J.; Boni, I.V.; Régnier, P.; Hajnsdorf, E. Hfq affects mRNA levels independently of degradation. BMC Mol. Biol. 2010, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, Y. Mechanism for coordinate regulation of rpoS by sRNA-sRNA interaction in Escherichia coli. RNA Biol. 2020, 17, 176–187. [Google Scholar] [CrossRef]
- Arce-Rodríguez, A.; Calles, B.; Nikel, P.I.; de Lorenzo, V. The RNA chaperone Hfq enables the environmental stress tolerance super-phenotype of Pseudomonas putida. Environ. Microbiol. 2016, 18, 3309–3326. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.; Li, F.; Zhang, J.; Wu, J.; Gong, Q.; Shi, Y. Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq. Nucleic Acids Res. 2015, 43, 2400–2411. [Google Scholar] [CrossRef][Green Version]
- Peng, Y.; Soper, T.J.; Woodson, S.A. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J. Mol. Biol. 2014, 426, 275–285. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Wu, J.; Gong, Q.; Shi, Y. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013, 41, 5938–5948. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, P.; Wang, Z.; Zhu, Q.; Xie, Z.; Mei, Y.; Liang, Y.; Chen, Z. Stress preadaptation and overexpression of rpoS and hfq genes increase stress resistance of Pseudomonas fluorescens ATCC13525. Microbiol. Res. 2021, 250, 126804. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993, 175, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Geiser, M.; Cèbe, R.; Drewello, D.; Schmitz, R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 2001, 31, 88–90. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Nejman, B.; Loś, J.M.; Łoś, M.; Węgrzyn, G.; Węgrzyn, A. Plasmids derived from lambdoid bacteriophages as models for studying replication of mobile genetic elements responsible for the production of Shiga toxins by pathogenic Escherichia coli strains. J. Mol. Microbiol. Biotechnol. 2009, 17, 211–220. [Google Scholar] [CrossRef]
- Kues, U.; Stahl, U. Replication of plasmids in gramnegative bacteria. Microbiol. Rev. 1989, 53, 491–516. [Google Scholar] [CrossRef]
- Bolivar, F.; Rodriguez, R.L.; Greene, P.J.; Betlach, M.C.; Heyneker, H.L.; Boyer, H.W.; Crosa, J.H.; Falkow, S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 1977, 2, 95–113. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory, Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Bloch, S.; Nejman-Faleńczyk, B.; Dydecka, A.; Łoś, J.M.; Felczykowska, A.; Węgrzyn, A.; Węgrzyn, G. Different expression patterns of genes from the exo-xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Φ24B following infection or prophage induction in Escherichia coli. PLoS ONE 2014, 9, e108233. [Google Scholar] [CrossRef]
- Roche. LightCycler Real-Time PCR Systems—Application Manual; Roche Diagnostics GmbH: Mannheim, Germany, 2009; p. 140. [Google Scholar]


| Plasmid | Replicon | Marker(s) | Copy Number/Cell |
|---|---|---|---|
| p27cmr | Lambdoid | cat (CmR) | Low (6–7) |
| pSC101 | pSC101 | tet (TetR) | Low (~5) |
| pBR322 | pMB1 (ColE1-like) | bla (ApR), tet (TetR) | Medium (20–40) |
| pBR332Δrom | pMB1 (ColE1-like) | bla (ApR), tet (TetR), cat (CmR) | High (~100) |
| Strain | Percent of Resistant Cells 1 | |
|---|---|---|
| 50 μg/mL Kanamycin | 233 μg/mL Kanamycin | |
| hfq+ | 100% | 0.012 ± 0.003% |
| Δhfq::kan | 100% | 49.64 ± 5.11% |
| ΔCTRhfq::kan | 100% | 1.26 ± 0.29% |
| Strain | Efficiency of Transformation (Transformants/μg DNA) 1 | |||
|---|---|---|---|---|
| p27cmr | pSC101 | pBR322 | pBR322Δrom | |
| hfq+ | 1.09 ± 0.42 × 105 | 1.72 ± 0.13 × 104 | 8.57 ± 0.61 × 104 | 2.58 ± 0.34 × 105 |
| Δhfq::kan | 1.21 ± 0.19 × 103 (*) | 2.03 ± 0.48 × 103 (*) | 1.53 ± 0.22 × 104 (*) | 5.60 ± 0.18 × 103 (*) |
| ΔCTRhfq::kan | 1.39 ± 0.25 × 104 (* #) | 3.11 ± 0.37 × 103 (*) | 3.04 ± 0.16 × 104 (*) | 6.51 ± 0.75 × 104 (* #) |
| Strain | Percent of Resistant Cells 1 | |
|---|---|---|
| 34 μg/mL Chloramphenicol | 204 μg/mL Chloramphenicol | |
| hfq+/p27cmr | 100% | 88.00 ± 5.66% |
| Δhfq::kan/p27cmr | 100% | 0.06 ± 0.02% |
| ΔCTRhfq::kan/p27cmr | 100% | 0.48 ± 0.17% |
| Strain | Percent of Resistant Cells 1 | |
|---|---|---|
| 12.5 μg/mL Tetracycline | 68 μg/mL Tetracycline | |
| hfq+/pSC101 | 100% | 17.69 ± 3.27% |
| Δhfq::kan/pSC101 | 100% | <0.0001% |
| ΔCTRhfq::kan/pSC101 | 100% | 38.88 ± 5.19% |
| Scheme 1 | Percent of Resistant Cells 1 | |
|---|---|---|
| 12.5 μg/mL Tetracycline | 68 μg/mL Tetracycline | |
| hfq+/pBR322 | 100% | 86.06 ± 10.44% |
| Δhfq::kan/pBR322 | 100% | 71.09 ± 12.03% |
| ΔCTRhfq::kan/pBR322 | 100% | 78.59 ± 16.66% |
| Strain | Percent of Resistant Cells 1 | |
|---|---|---|
| 50 μg/mL Ampicillin | 5 mg/mL Ampicillin | |
| hfq+/pBR322 | 100% | 69.76 ± 8.07% |
| Δhfq::kan/pBR322 | 100% | 9.00 ± 1.82% |
| ΔCTRhfq::kan/pBR322 | 100% | 87.90 ± 10.29% |
| Strain | OD600 of Bacterial Culture | pBR322 Copy Number 1 |
|---|---|---|
| hfq+/pBR322 | 0.3 | 18.60 ± 1.84 |
| 0.8 | 37.98 ± 1.11 | |
| 1.4 | 21.31 ± 1.76 | |
| Δhfq::kan/pBR322 | 0.3 | 23.30 ± 1.02 |
| 0.8 | 41.09 ± 2.37 | |
| 1.4 | 27.80 ± 1.90 | |
| ΔCTRhfq::kan/pBR322 | 0.3 | 21.77 ± 2.16 |
| 0.8 | 43.07 ± 2.94 | |
| 1.4 | 27.03 ± 4.08 |
| Primer Name | Sequence | Description |
|---|---|---|
| Primer 1 | 5′-CGG GGA TCC GTC GAC CTG CAG TAT GGC TAA GGG GCA ATC TTT AC | Forward primer containing a sequence complementary to the 5′-region of hfq and a specific sequence of pKD13 |
| Primer 2 | 5′-AAG TTC CTA TAC TTT CTA GAG AAT AGG AAC TTC GAT TAT TCG GTT TCT TCG CTG TCC | Reverse primer containing a sequence complementary to the 5′-region of hfq and a specific sequence of pKD13 |
| Primer 3 | 5′-ACG CGA TTT CTA CTG TTG TCC CGT AAT CGA AGT TCC TAT TCT CTA GA | Forward primer containing a sequence complementary to the distal part of NTR of hfq on pKD13 and the termination sequence |
| Primer 4 | 5′-TCA AAA GCG CTC TGA AGT TCC TAT ACT TTC TAG AGA ATA GGA ACT TCG ATT ACG | Reverse primer containing a sequence complementary to the distal part of NTR of hfq on pKD13 and the termination sequence |
| Primer 5 | 5′-TCA GAA TCG AAA GGT TCA AAG TAC AAA TAA GCA TAT AAG GAA AAG AGA GAA TGG CTA AGG GGC AAT CTT T | Forward primer containing a sequence complementary to the5′-region of hfq and overhang complementary to the hfq region on E. coli MG1655 genome |
| Primer 6 | 5′-CGG GGA ACG CAG GAT CGC TGG CTC CCC GTG TAA AAA AAC AGC CCG AAA CCT GTG TAG GCT GGA GCT GCT TC | Reverse primer containing a sequence complementary to the 5′-region of hfq and overhang complementary to the hfq region on E. coli MG1655 genome |
| Primer 7 | 5′-TCA GAA TCG AAA GGT TCA AAG TAC AAA TAA GCA TAT AAG GAA AAG AGA GAA TGA TTG AAC AAG ATG GAT TG | Forward primer containing a sequence complementary to the 5′-region of kan and an overhang complementary to the hfq region on E. coli MG1655 genome |
| Primer 8 | 5′-CAC ATG CAG CTC CCG GAG ACG | Forward primer for amplification of pBR322 without rom |
| Primer 9 | 5′-TGA TGC CTC CGT GTA AGG GGG | Reverse primer for amplification of pBR322 without rom |
| Primer 10 | 5′-TTA AAA AAA TTA CGC CCC GCC | Forward primer for amplification of the cat gene |
| Primer 11 | 5′-ATG GAG AAA AAA ATC TCT GGA | Reverse primer for amplification of the cat gene |
| Primer 12 | 5′-ACC GCA GAA CGT ATC AAG CA | Forward primer for amplification of the mreB gene |
| Primer 13 | 5′-GG TAA AAC CGC GTG GAA CAC | Reverse primer for amplification of the mreB gene |
| Primer 14 | 5′-CTC ATC GTC ATC CTC GGC AC | Forward primer for amplification of the tet gene |
| Primer 15 | 5′-TAG CAG CAC GCC ATA GTG AC | Forward primer for amplification of the tet gene |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaffke, L.; Kubiak, K.; Cyske, Z.; Węgrzyn, G. Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics. Int. J. Mol. Sci. 2021, 22, 8886. https://doi.org/10.3390/ijms22168886
Gaffke L, Kubiak K, Cyske Z, Węgrzyn G. Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics. International Journal of Molecular Sciences. 2021; 22(16):8886. https://doi.org/10.3390/ijms22168886
Chicago/Turabian StyleGaffke, Lidia, Krzysztof Kubiak, Zuzanna Cyske, and Grzegorz Węgrzyn. 2021. "Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics" International Journal of Molecular Sciences 22, no. 16: 8886. https://doi.org/10.3390/ijms22168886
APA StyleGaffke, L., Kubiak, K., Cyske, Z., & Węgrzyn, G. (2021). Differential Chromosome- and Plasmid-Borne Resistance of Escherichia coli hfq Mutants to High Concentrations of Various Antibiotics. International Journal of Molecular Sciences, 22(16), 8886. https://doi.org/10.3390/ijms22168886








