Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation
Abstract
:1. Introduction
2. Results
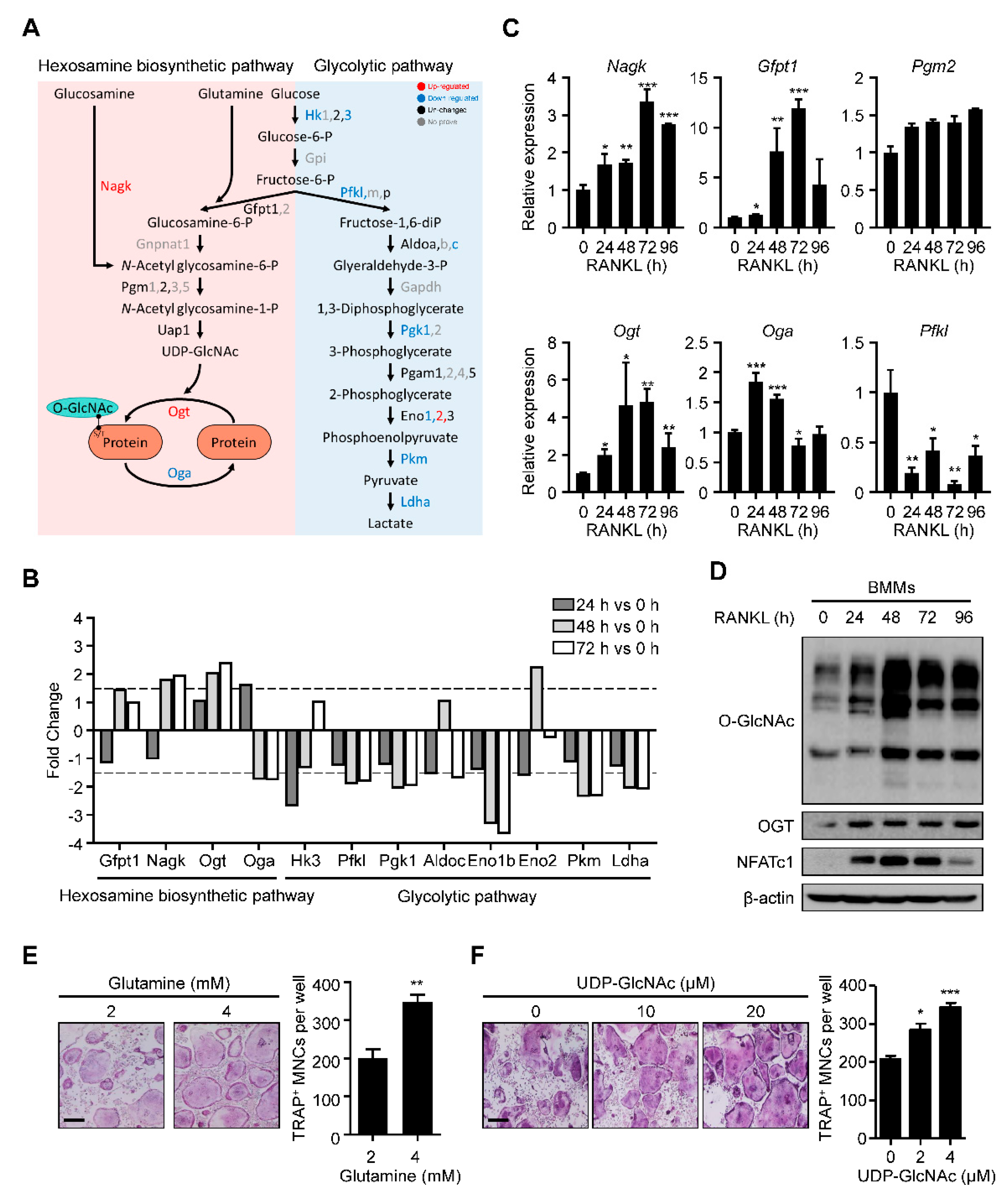
2.1. HBP Is Activated and O-GlcNAcylation Is Increased during RANKL-Mediated Osteoclast Differentiation
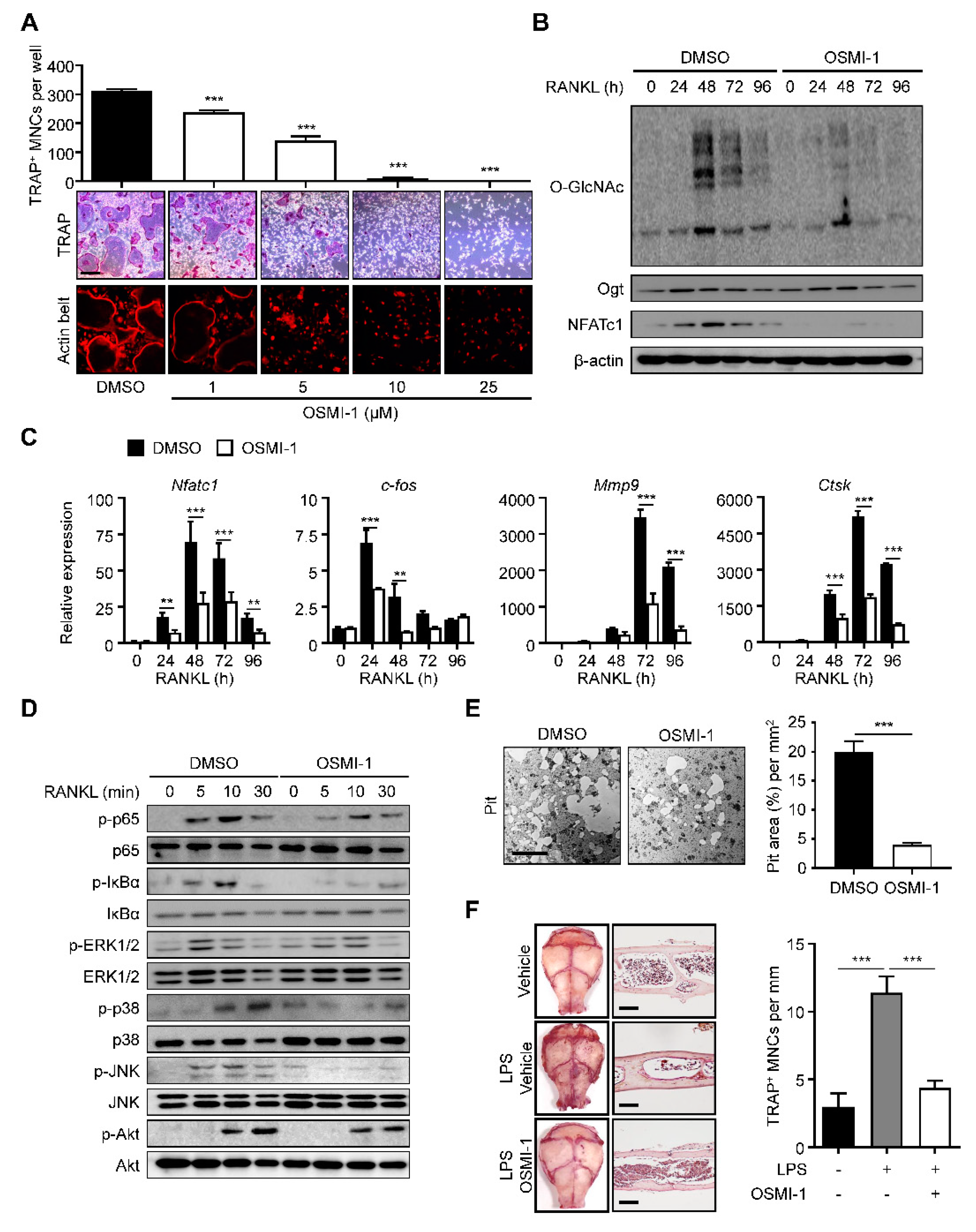
2.2. Inhibition of O-GlcNAcylation Interferes with Osteoclastogenesis
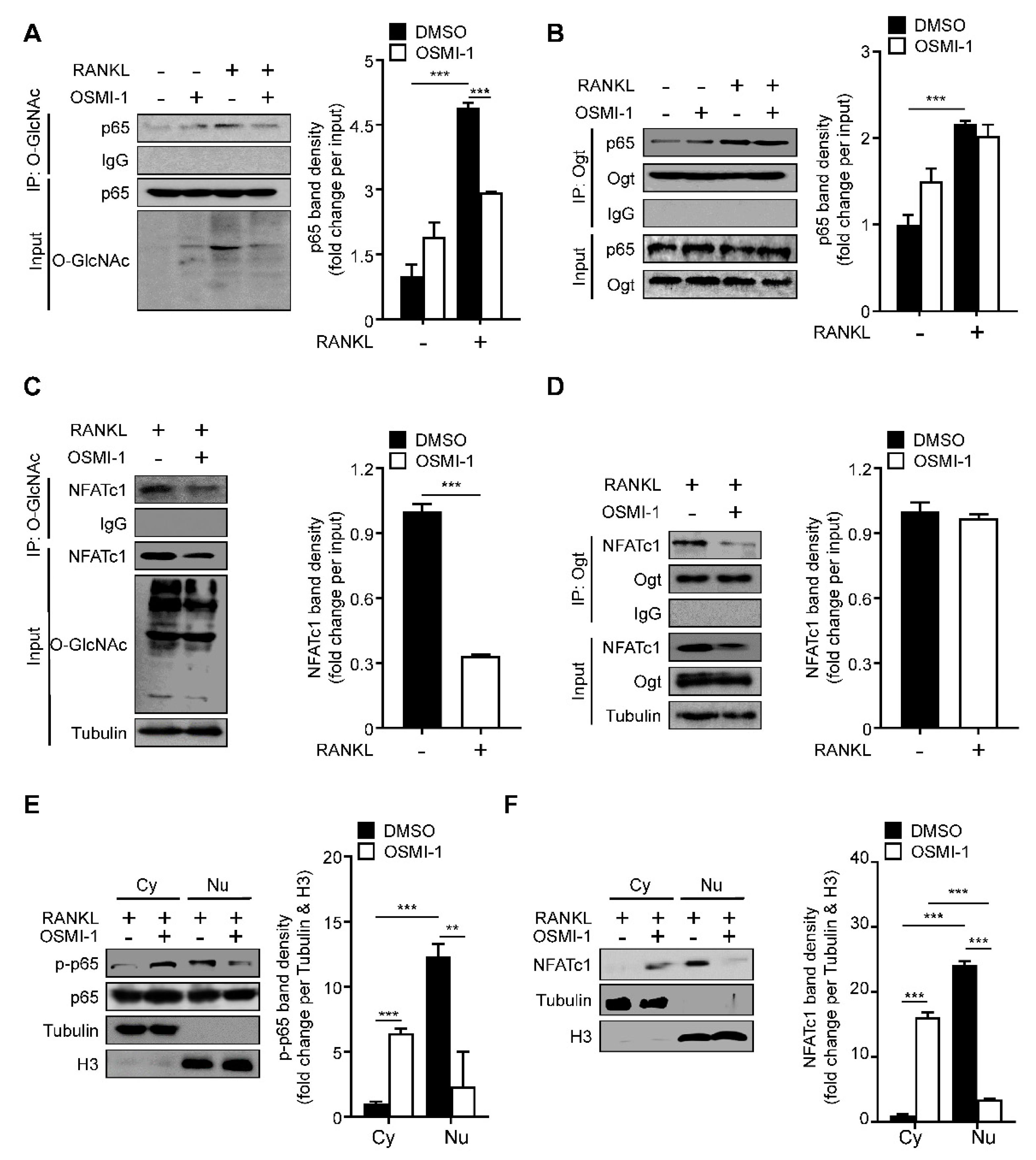
2.3. RANKL Stimulation Induces O-GlcNAcylation of NF-κB p65 and NFATc1
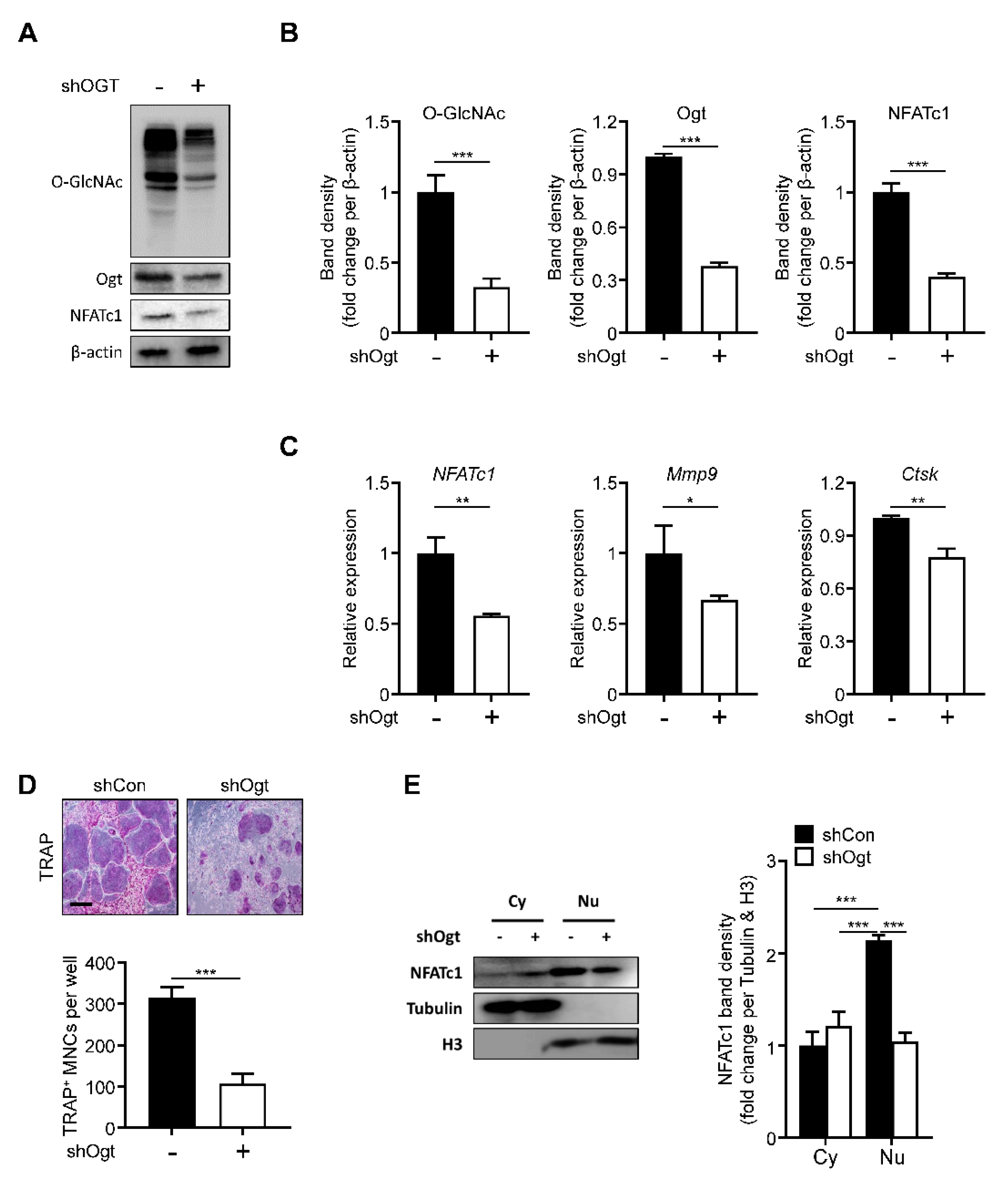
2.4. Ogt Knockdown Mimics the Effects of OSMI-1
2.5. Ogt Inhibition Shows a Synergistic Effect with Bone Target Therapy on Osteoclastogenesis
3. Discussion
4. Materials and Methods
4.1. Isolation and Osteoclast Differentiation
4.2. TRAP Staining and Actin Belt Formation Assay
4.3. Cell Viability Assay
4.4. Bone Resorption Assay
4.5. Lentiviral-Mediated Gene Transduction
4.6. RNA Extraction and RT-qPCR
4.7. Western Blot
4.8. Immunoprecipitation
4.9. Cytoplasmic Extract and Nuclear Extraction
4.10. Calvarial Bone Destruction Assay
4.11. Transcriptome Analysis
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lothrop, A.P.; Torres, M.P.; Fuchs, S.M. Deciphering post-translational modification codes. FEBS Lett. 2013, 587, 1247–1257. [Google Scholar] [CrossRef] [Green Version]
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022. [Google Scholar] [CrossRef]
- Chiaradonna, F.; Ricciardiello, F.; Palorini, R. The Nutrient-Sensing Hexosamine Biosynthetic Pathway as the Hub of Cancer Metabolic Rewiring. Cells 2018, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Rossetti, L. Perspective: Hexosamines and nutrient sensing. Endocrinology 2000, 141, 1922–1925. [Google Scholar] [CrossRef]
- Yuzwa, S.A.; Shan, X.; Macauley, M.S.; Clark, T.; Skorobogatko, Y.; Vosseller, K.; Vocadlo, D.J. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 2012, 8, 393–399. [Google Scholar] [CrossRef]
- Ruan, H.B.; Singh, J.P.; Li, M.D.; Wu, J.; Yang, X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 2013, 24, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Bond, M.R.; Hanover, J.A. O-GlcNAc Cycling: A Link Between Metabolism and Chronic Disease. Annu. Rev. Nutr. 2013, 33, 205–229. [Google Scholar] [CrossRef]
- Slawson, C.; Hart, G.W. O-GlcNAc signalling: Implications for cancer cell biology. Nat. Rev. Cancer 2011, 11, 678–684. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef]
- Wells, L.; Gao, Y.; Mahoney, J.A.; Vosseller, K.; Chen, C.; Rosen, A.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: Further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002, 277, 1755–1761. [Google Scholar] [CrossRef] [Green Version]
- Sohn, K.C.; Lee, E.J.; Shin, J.M.; Lim, E.H.; No, Y.; Lee, J.Y.; Yoon, T.Y.; Lee, Y.H.; Im, M.; Lee, Y.; et al. Regulation of keratinocyte differentiation by O-GlcNAcylation. J. Dermatol. Sci. 2014, 75, 10–15. [Google Scholar] [CrossRef]
- Berrabah, W.; Aumercier, P.; Gheeraert, C.; Dehondt, H.; Bouchaert, E.; Alexandre, J.; Ploton, M.; Mazuy, C.; Caron, S.; Tailleux, A.; et al. Glucose sensing O-GlcNAcylation pathway regulates the nuclear bile acid receptor farnesoid X receptor (FXR). Hepatology 2014, 59, 2022–2033. [Google Scholar] [CrossRef]
- Golks, A.; Tran, T.T.; Goetschy, J.F.; Guerini, D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007, 26, 4368–4379. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, P.; Clark, P.M.; Mason, D.E.; Peters, E.C.; Hsieh-Wilson, L.C.; Baltimore, D. Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 2013, 6, ra75. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Molina, H.; Huang, H.; Zhang, Y.Y.; Liu, M.; Qian, S.W.; Slawson, C.; Dias, W.B.; Pandey, A.; Hart, G.W.; et al. O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein beta: Role during adipocyte differentiation. J. Biol. Chem. 2009, 284, 19248–19254. [Google Scholar] [CrossRef] [Green Version]
- Vosseller, K.; Wells, L.; Lane, M.D.; Hart, G.W. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 5313–5318. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Schwarz, T.L. O-GlcNAc Transferase Is Essential for Sensory Neuron Survival and Maintenance. J. Neurosci. 2017, 37, 2125–2136. [Google Scholar] [CrossRef] [Green Version]
- Rexach, J.E.; Clark, P.M.; Mason, D.E.; Neve, R.L.; Peters, E.C.; Hsieh-Wilson, L.C. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 2012, 8, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Swamy, M.; Pathak, S.; Grzes, K.M.; Damerow, S.; Sinclair, L.V.; van Aalten, D.M.; Cantrell, D.A. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 2016, 17, 712–720. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.H.; Park, S.Y.; Nam, H.W.; Kim, D.H.; Kang, J.G.; Kang, E.S.; Kim, Y.S.; Lee, H.C.; Kim, K.S.; Cho, J.W. NF kappa B activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 17345–17350. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Chalkley, R.J.; Vosseller, K. Hyper-O-GlcNAcylation activates nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappaB) signaling through interplay with phosphorylation and acetylation. J. Biol. Chem. 2017, 292, 9150–9163. [Google Scholar] [CrossRef] [Green Version]
- Dentin, R.; Hedrick, S.; Xie, J.; Yates, J., 3rd; Montminy, M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science 2008, 319, 1402–1405. [Google Scholar] [CrossRef]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Suda, T.; Takahashi, N.; Udagawa, N.; Jimi, E.; Gillespie, M.T.; Martin, T.J. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999, 20, 345–357. [Google Scholar] [CrossRef]
- Walsh, M.C.; Kim, N.; Kadono, Y.; Rho, J.; Lee, S.Y.; Lorenzo, J.; Choi, Y. Osteoimmunology: Interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 2006, 24, 33–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Kamemura, K. Global increase in O-linked N-acetylglucosamine modification promotes osteoblast differentiation. Exp. Cell Res. 2015, 338, 194–202. [Google Scholar] [CrossRef]
- Nagel, A.K.; Ball, L.E. O-GlcNAc modification of the runt-related transcription factor 2 (Runx2) links osteogenesis and nutrient metabolism in bone marrow mesenchymal stem cells. Mol. Cell. Proteom. 2014, 13, 3381–3395. [Google Scholar] [CrossRef] [Green Version]
- Nagel, A.K.; Schilling, M.; Comte-Walters, S.; Berkaw, M.N.; Ball, L.E. Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation. Mol. Cell. Proteom. 2013, 12, 945–955. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kim, Y.H.; Song, M.; An, S.H.; Byun, H.Y.; Heo, K.; Lim, S.; Oh, Y.S.; Ryu, S.H.; Suh, P.G. O-GlcNAc modification modulates the expression of osteocalcin via OSE2 and Runx2. Biochem. Biophys. Res. Commun. 2007, 362, 325–329. [Google Scholar] [CrossRef]
- Gu, H.; Song, M.; Boonanantanasarn, K.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Baek, J.H. Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells. Int. J. Mol. Sci. 2018, 19, 202. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Qi, M.; Wang, Y.; Feng, X.; Liu, H. Zoledronate and high glucose levels influence osteoclast differentiation and bone absorption via the AMPK pathway. Biochem. Biophys. Res. Commun. 2018, 505, 1195–1202. [Google Scholar] [CrossRef]
- Xu, J.; Yue, F.; Wang, J.; Chen, L.; Qi, W. High glucose inhibits receptor activator of nuclear factorkappaB ligand-induced osteoclast differentiation via downregulation of vATPase V0 subunit d2 and dendritic cellspecific transmembrane protein. Mol. Med. Rep. 2015, 11, 865–870. [Google Scholar] [CrossRef] [Green Version]
- Wittrant, Y.; Gorin, Y.; Woodruff, K.; Horn, D.; Abboud, H.E.; Mohan, S.; Abboud-Werner, S.L. High d(+)glucose concentration inhibits RANKL-induced osteoclastogenesis. Bone 2008, 42, 1122–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.M.; Jeong, D.; Kang, H.K.; Jung, S.Y.; Kang, S.S.; Min, B.M. Osteoclast precursors display dynamic metabolic shifts toward accelerated glucose metabolism at an early stage of RANKL-stimulated osteoclast differentiation. Cell. Physiol. Biochem. 2007, 20, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ren, Y.; Cao, Y.; Huang, H.; Wu, Q.; Li, W.; Wu, S.; Zhang, J. Discovery of a Low Toxicity O-GlcNAc Transferase (OGT) Inhibitor by Structure-based Virtual Screening of Natural Products. Sci. Rep. 2017, 7, 12334. [Google Scholar] [CrossRef] [Green Version]
- Lyles, K.W.; Colon-Emeric, C.S.; Magaziner, J.S.; Adachi, J.D.; Pieper, C.F.; Mautalen, C.; Hyldstrup, L.; Recknor, C.; Nordsletten, L.; Moore, K.A.; et al. Zoledronic acid and clinical fractures and mortality after hip fracture. N. Engl. J. Med. 2007, 357, 1799–1809. [Google Scholar] [CrossRef] [Green Version]
- McColgan, N.M.; Feeley, M.N.; Woodward, A.M.; Guindolet, D.; Argueso, P. The O-GlcNAc modification promotes terminal differentiation of human corneal epithelial cells. Glycobiology 2020, 30, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.J.; Lin, T.; Hsieh, P.C.; Liao, M.C.; Shin, S.J. Suppression of Glutamine:fructose-6-phosphate amidotransferase-1 inhibits adipogenesis in 3T3-L1 adipocytes. J. Cell. Physiol. 2012, 227, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Andres-Bergos, J.; Tardio, L.; Larranaga-Vera, A.; Gomez, R.; Herrero-Beaumont, G.; Largo, R. The increase in O-linked N-acetylglucosamine protein modification stimulates chondrogenic differentiation both in vitro and in vivo. J. Biol. Chem. 2012, 287, 33615–33628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihara, K.; Takahashi, I.; Tsuchiya, Y.; Hasegawa, M.; Kamemura, K. Characteristic increase in nucleocytoplasmic protein glycosylation by O-GlcNAc in 3T3-L1 adipocyte differentiation. Biochem. Biophys. Res. Commun. 2010, 398, 489–494. [Google Scholar] [CrossRef]
- Zhang, Z.; Parker, M.P.; Graw, S.; Novikova, L.V.; Fedosyuk, H.; Fontes, J.D.; Koestler, D.C.; Peterson, K.R.; Slawson, C. O-GlcNAc homeostasis contributes to cell fate decisions during hematopoiesis. J. Biol. Chem. 2019, 294, 1363–1379. [Google Scholar] [CrossRef] [Green Version]
- Andres, L.M.; Blong, I.W.; Evans, A.C.; Rumachik, N.G.; Yamaguchi, T.; Pham, N.D.; Thompson, P.; Kohler, J.J.; Bertozzi, C.R. Chemical Modulation of Protein O-GlcNAcylation via OGT Inhibition Promotes Human Neural Cell Differentiation. ACS Chem. Biol. 2017, 12, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Mizofuchi, H.; Kobayashi, Y.; Tsuzuki, G.; Yamamoto, M.; Wada, S.; Kamemura, K. Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 24–32. [Google Scholar] [CrossRef]
- Zhang, D.; Cai, Y.; Chen, M.; Gao, L.; Shen, Y.; Huang, Z. OGT-mediated O-GlcNAcylation promotes NF-kappaB activation and inflammation in acute pancreatitis. Inflamm. Res. 2015, 64, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Allison, D.F.; Wamsley, J.J.; Kumar, M.; Li, D.; Gray, L.G.; Hart, G.W.; Jones, D.R.; Mayo, M.W. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-kappaB acetylation and transcription. Proc. Natl. Acad. Sci. USA 2012, 109, 16888–16893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Kim, K.; Youn, B.U.; Jin, H.M.; Kim, J.Y.; Moon, J.B.; Ko, A.; Seo, S.B.; Lee, K.Y.; Kim, N. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 2011, 436, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Horimoto, Y.; Oyama, M.; Nakatani, S.; Kobata, K.; Tamura, M.; Arata, Y.; Hatanaka, T. Osteoclast Differentiation Is Suppressed by Increased O-GlcNAcylation Due to Thiamet G Treatment. Biol. Pharm. Bull. 2020, 43, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Sugimoto, A.; Imazato, N.; Tamura, M.; Nakatani, S.; Kobata, K.; Arata, Y. Glucosamine Suppresses Osteoclast Differentiation through the Modulation of Glycosylation Including O-GlcNAcylation. Biol. Pharm. Bull. 2017, 40, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, T.; Nagasaka, M.; Shimizu, M.; Tamura, M.; Arata, Y. N-acetylglucosamine suppresses osteoclastogenesis in part through the promotion of O-GlcNAcylation. Bone Rep. 2016, 5, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Tan, E.P.; VandenHull, N.J.; Peterson, K.R.; Slawson, C. O-GIcNAcase expression is sensitive to changes in O-GIcNAc homeostasis. Front. Endocrinol. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Khidekel, N.; Ficarro, S.B.; Clark, P.M.; Bryan, M.C.; Swaney, D.L.; Rexach, J.E.; Sun, Y.E.; Coon, J.J.; Peters, E.C.; Hsieh-Wilson, L.C. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007, 3, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar] [CrossRef] [Green Version]
- Cummings, S.R.; San Martin, J.; McClung, M.R.; Siris, E.S.; Eastell, R.; Reid, I.R.; Delmas, P.; Zoog, H.B.; Austin, M.; Wang, A.; et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N. Engl. J. Med. 2009, 361, 756–765. [Google Scholar] [CrossRef] [Green Version]
- Esposito, M.; Ganesan, S.; Kang, Y. Emerging strategies for treating metastasis. Nat. Cancer 2021, 2, 258–270. [Google Scholar] [CrossRef]
- Wu, D.; Jin, J.; Qiu, Z.; Liu, D.; Luo, H. Functional Analysis of O-GlcNAcylation in Cancer Metastasis. Front. Oncol. 2020, 10, 585288. [Google Scholar] [CrossRef]
- Mercatali, L.; La Manna, F.; Miserocchi, G.; Liverani, C.; De Vita, A.; Spadazzi, C.; Bongiovanni, A.; Recine, F.; Amadori, D.; Ghetti, M.; et al. Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition. Int. J. Mol. Sci. 2017, 18, 1655. [Google Scholar] [CrossRef] [Green Version]
- Mercatali, L.; Spadazzi, C.; Miserocchi, G.; Liverani, C.; De Vita, A.; Bongiovanni, A.; Recine, F.; Amadori, D.; Ibrahim, T. The Effect of Everolimus in an In Vitro Model of Triple Negative Breast Cancer and Osteoclasts. Int. J. Mol. Sci. 2016, 17, 1824. [Google Scholar] [CrossRef] [Green Version]

| Gene | Primer Sequences | |
|---|---|---|
| c-fos | Forward Reverse | 5′-ATCCGAAGGGAACGGAATAA-3′ 5′-TGGGCTGCCAAAATAAACTC-3′ |
| Ctsk | Forward Reverse | 5′-AGGCATTGACTCTGAAGATGCT-3′ 5′-TCCCCACAGGAATCTCTCTG-3′ |
| Gfpt1 | Forward Reverse | 5′-GGAAAAGTTAAGGCACTGGATG-3′ 5′-GGGGGTGACTATTGACAGGA-3′ |
| Mmp9 | Forward Reverse | 5′-TGTCATCCAGTTTGGTGTCG-3′ 5′-AATGGGCATCTCCCTGAAC-3′ |
| Nagk | Forward Reverse | 5′-TTGATTCCATCGACAACCTG-3′ 5′-CTGGCAAAATCCAGCAAACT-3′ |
| NFATc1 | Forward Reverse | 5′-GCCTTTTGCGAGCAGTATCT-3′ 5′-TCATAGTGAGCCCTGTGGTG-3′ |
| Oga | Forward Reverse | 5′-CAGTACCTGGGAGAGCCAGA-3′ 5′-GTCCAAAGCACCTCAATTCC-3′ |
| Ogt | Forward Reverse | 5′-CATGCAGCTCTGGAGACAAG-3′ 5′-TGTCGATAATGCTCGATTGC-3′ |
| Pfkl | Forward Reverse | 5′-AAGAAGTAGGCTGGCACGAC-3′ 5′-GCAGGGCGTGAATACCATAG-3′ |
| Pgm2 | Forward Reverse | 5′-AACAGCTCATTGCTGGAGGT-3′ 5′-GCAAAATCCCTGTGTGGTCT-3′ |
| β-actin | Forward Reverse | 5′-AAGTGTGACGTTGACATCCG-3′ 5′-GATCCACATCTGCTGGAAGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.J.; Kim, H.S.; Lee, S.; Min, K.Y.; Choi, W.S.; You, J.S. Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation. Int. J. Mol. Sci. 2021, 22, 8888. https://doi.org/10.3390/ijms22168888
Kim MJ, Kim HS, Lee S, Min KY, Choi WS, You JS. Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation. International Journal of Molecular Sciences. 2021; 22(16):8888. https://doi.org/10.3390/ijms22168888
Chicago/Turabian StyleKim, Myoung Jun, Hyuk Soon Kim, Sangyong Lee, Keun Young Min, Wahn Soo Choi, and Jueng Soo You. 2021. "Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation" International Journal of Molecular Sciences 22, no. 16: 8888. https://doi.org/10.3390/ijms22168888
APA StyleKim, M. J., Kim, H. S., Lee, S., Min, K. Y., Choi, W. S., & You, J. S. (2021). Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation. International Journal of Molecular Sciences, 22(16), 8888. https://doi.org/10.3390/ijms22168888







