Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus
Abstract
:1. Introduction
2. Results
2.1. Bacterial Isolation and Genome-Based Identification
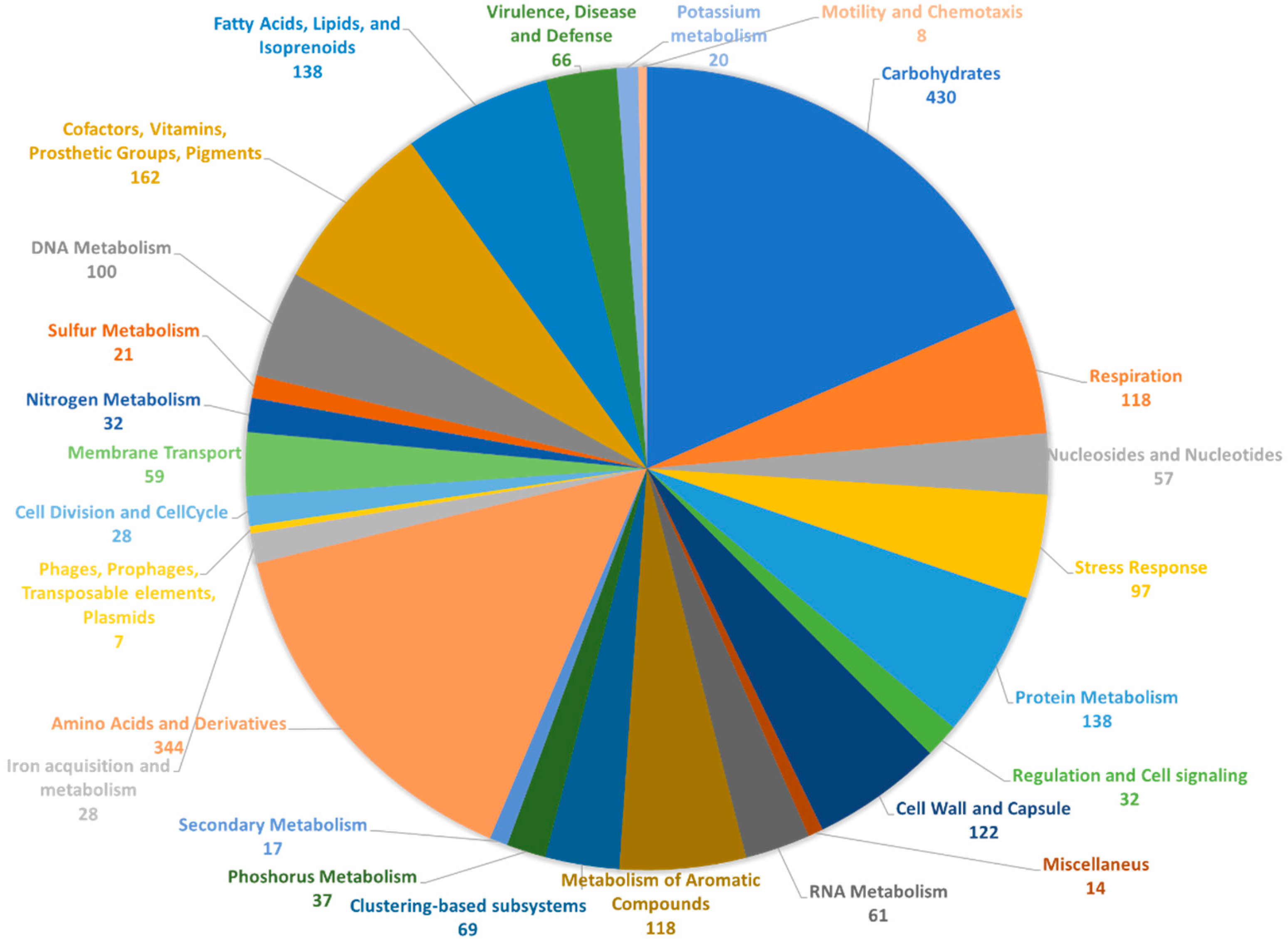
2.2. Genome Annotation and Biosynthetic Potential Analysis of Rhodococcus sp. I2R
2.3. OSMAC-Based Cultivation and Bioactivity Profiling
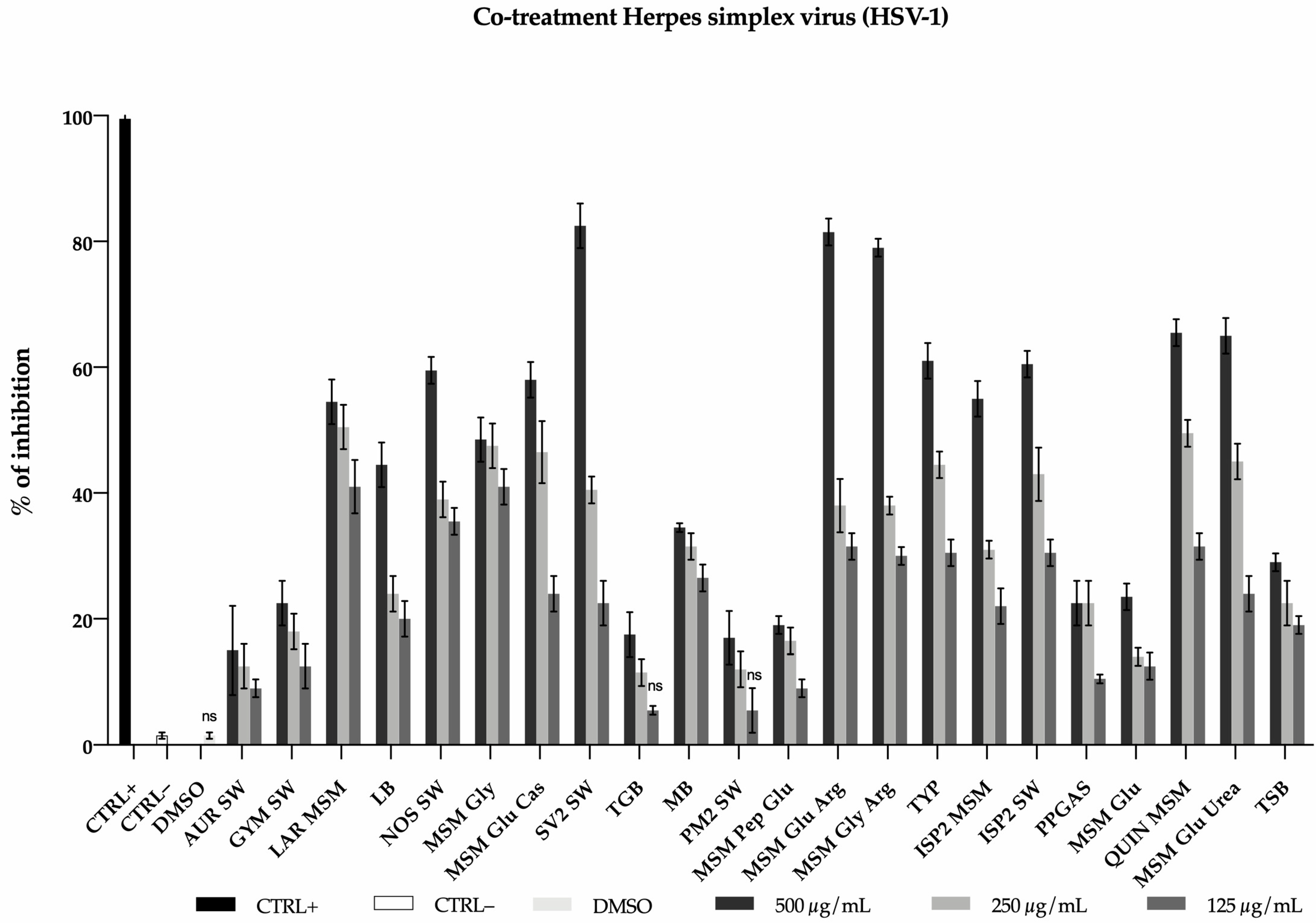
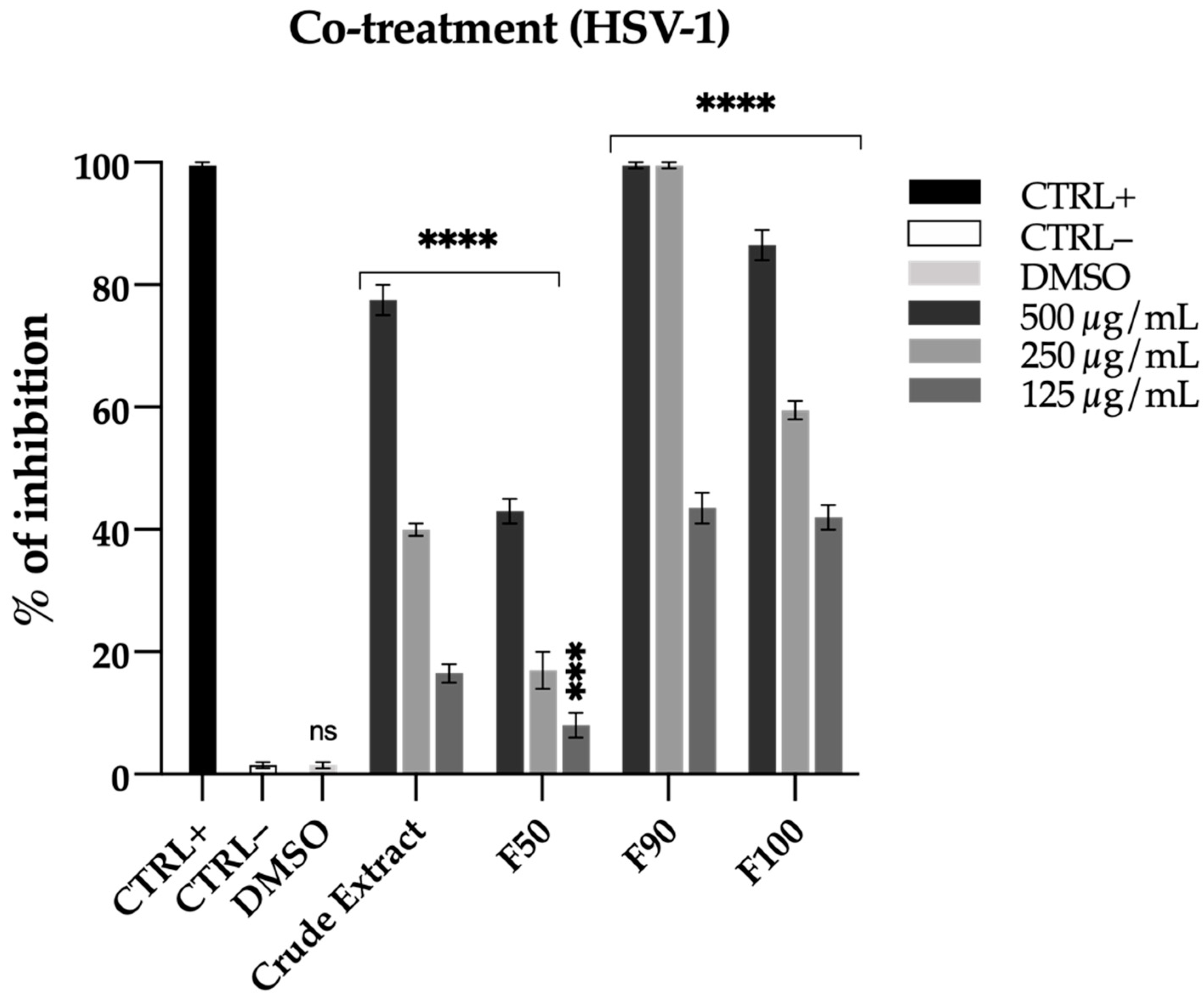
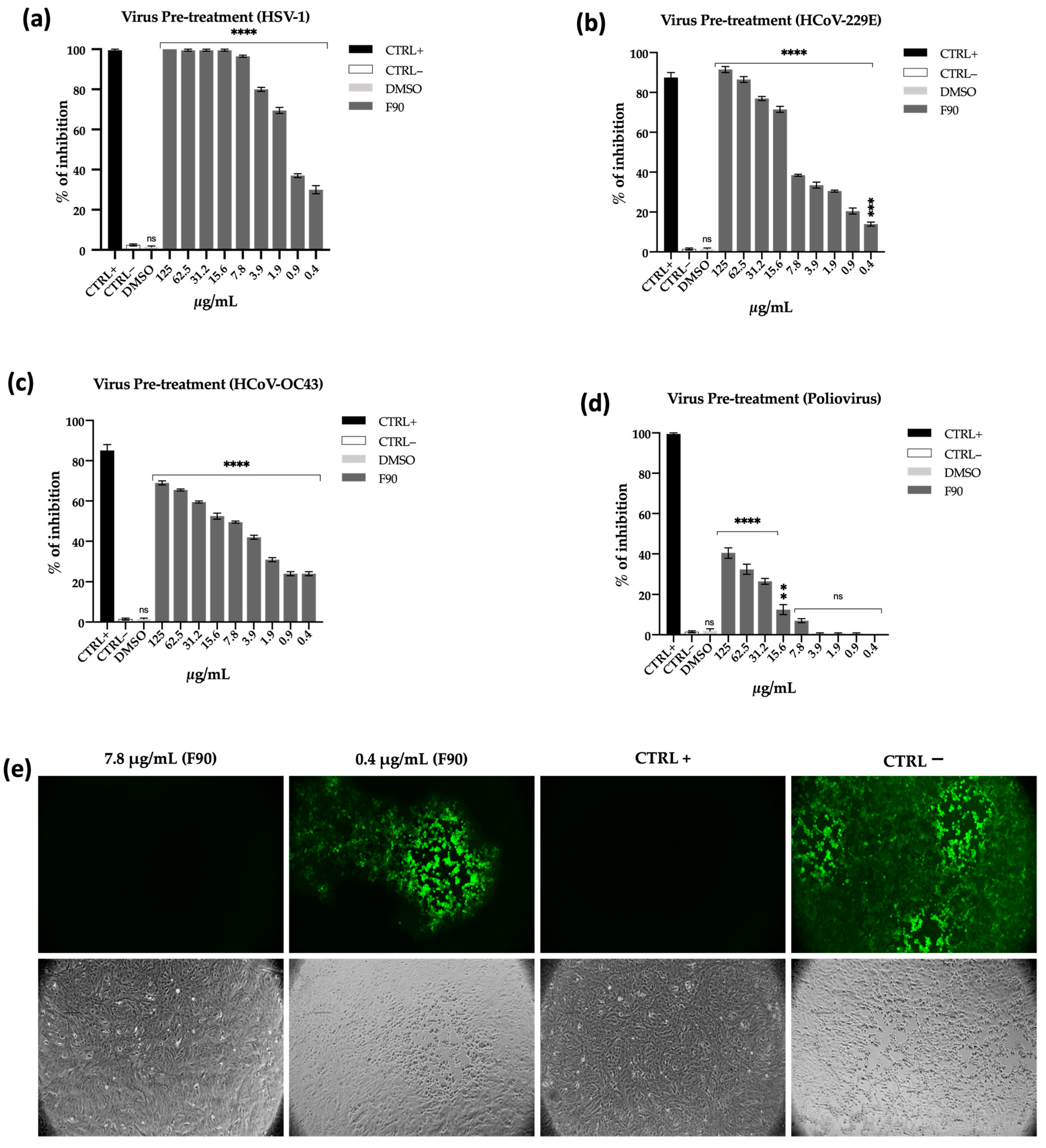
2.3.1. Antiviral Activity
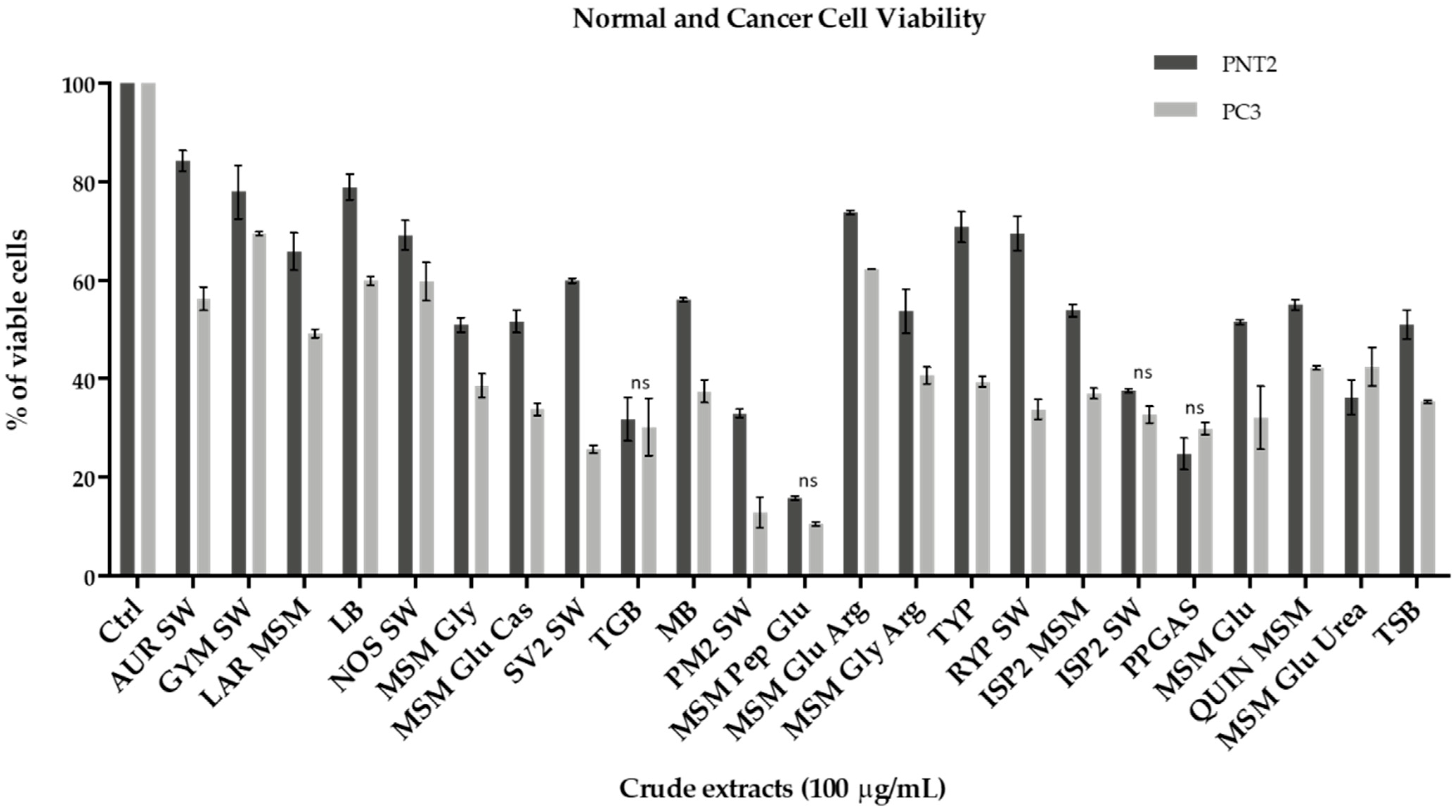
2.3.2. Anticancer Activity
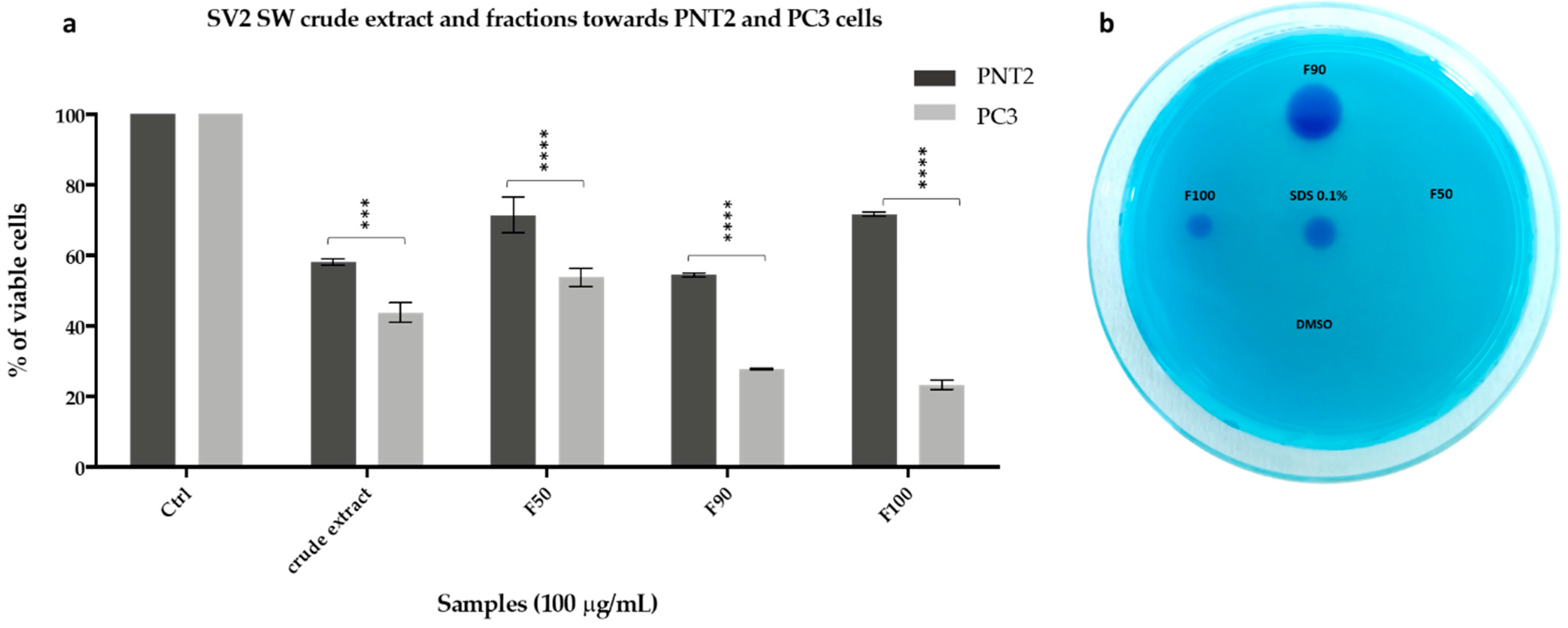
2.3.3. Biosurfactant Activity
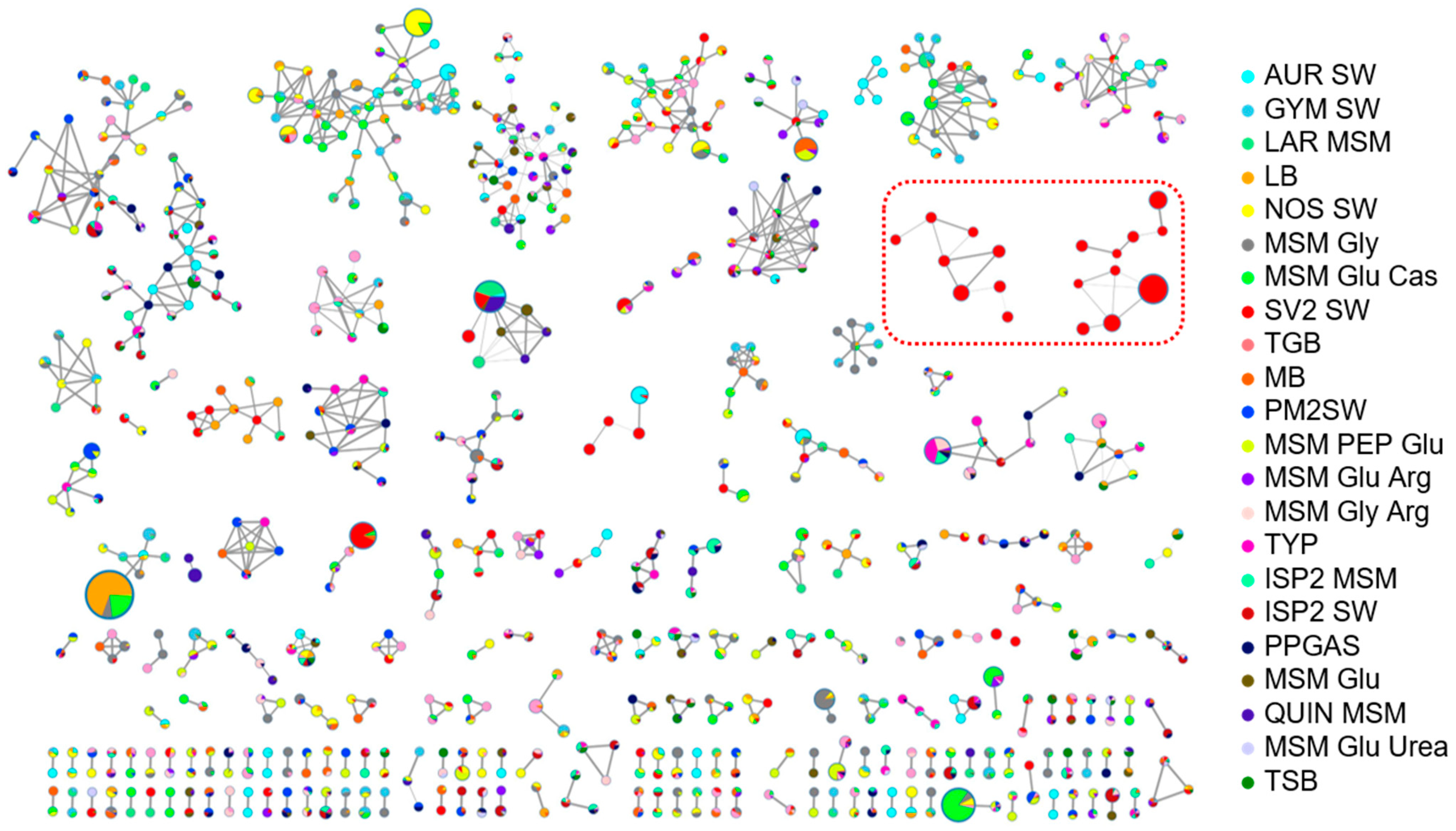
2.4. Molecular Networking Analysis of Rhodococcus sp. I2R Metabolism under Different Culture Conditions
2.5. Bioassay-Guided Fractionation of the SV2 SW Crude Extract
2.5.1. Antiviral Activity Validation
2.5.2. Antiproliferative and Biosurfactant Activity Validation
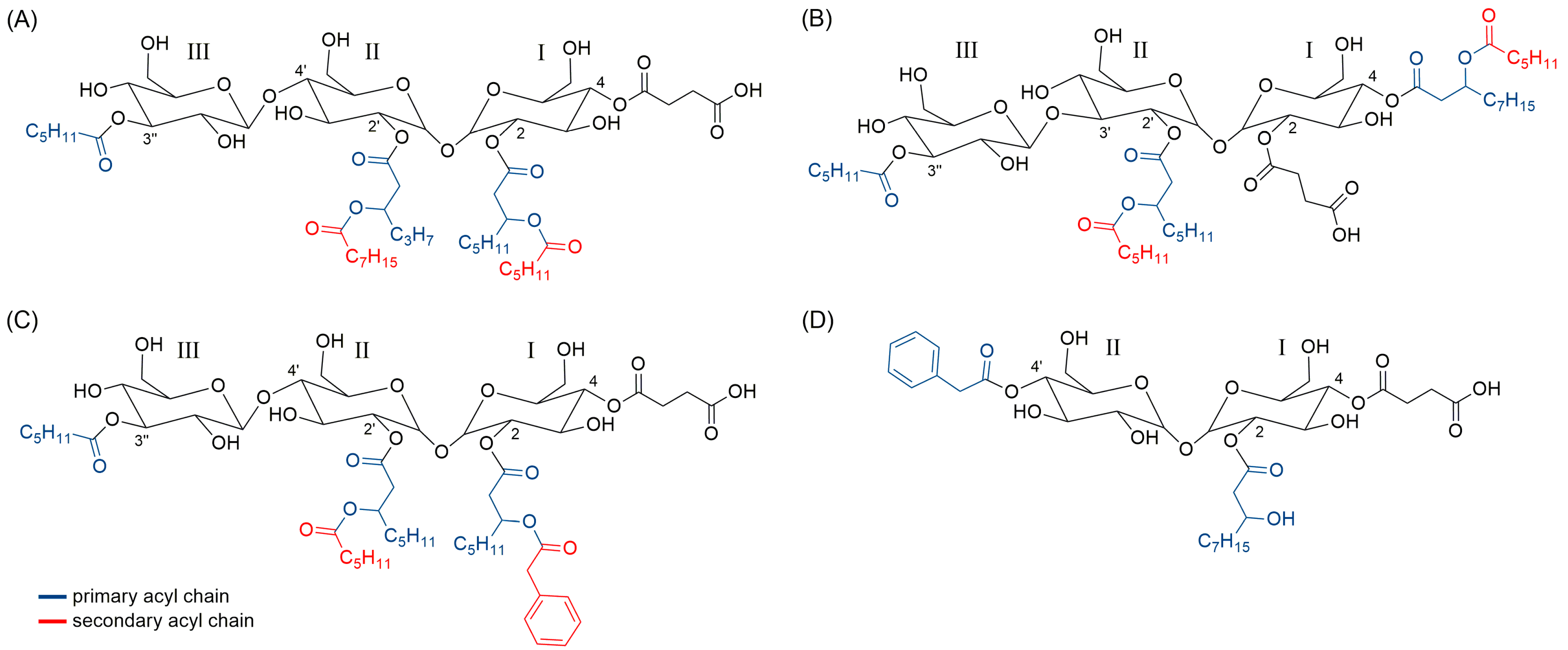
2.6. Structure Prediction of Novel Succinic Saccharide Esters
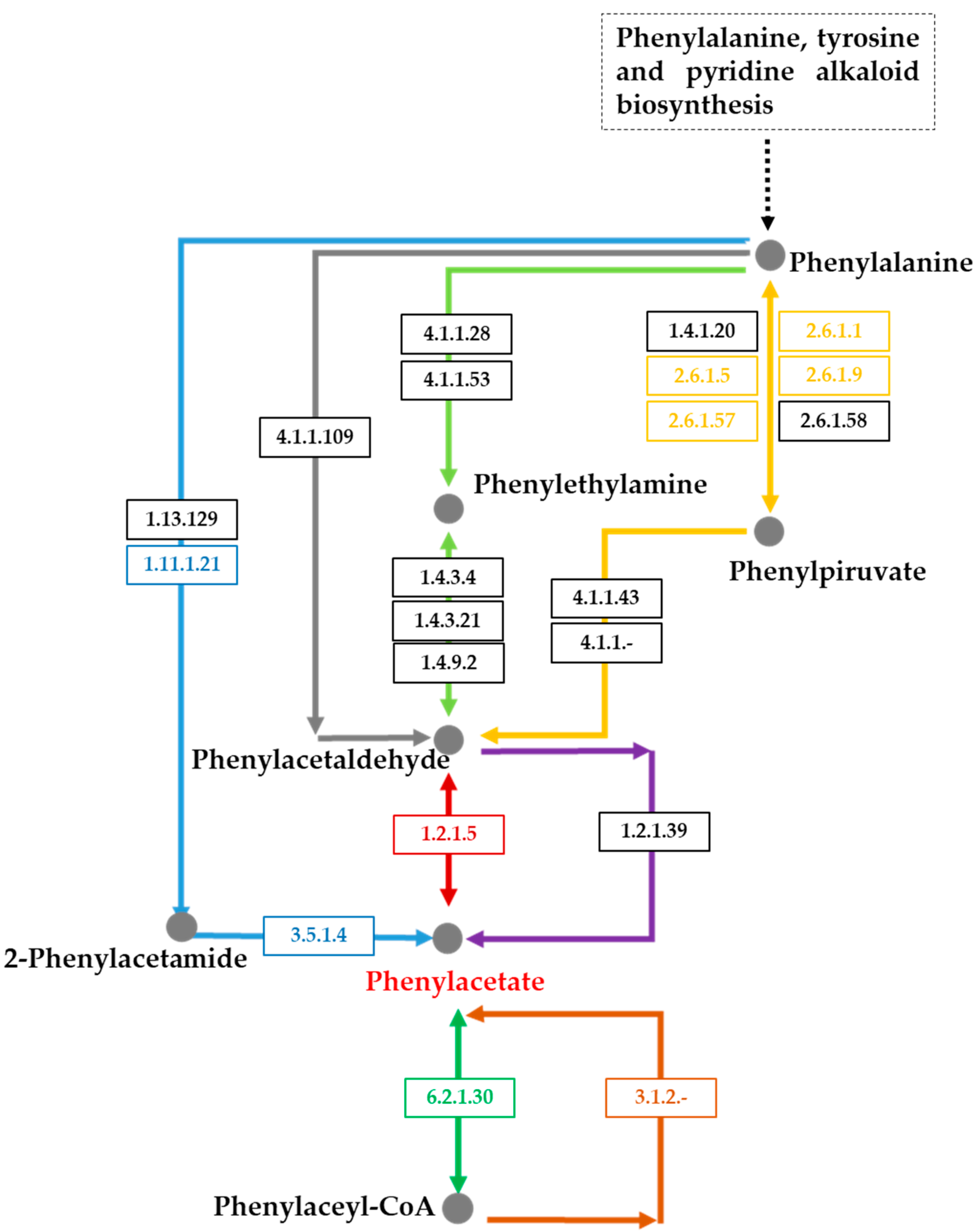
2.7. Identification of Putative Genes Involved in Succinic Saccharide Esters Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Isolation, Identification, and Genome Sequencing of Strain I2R
4.2. Genome Annotation
4.3. OSMAC Approach and Growth Media
4.4. Preparation of Crude Extracts
4.5. Bioactivity Evaluation by Functional Assays
4.5.1. Antiviral Assays
Virus, Cell Culture and Treatment
HSV-1 Co-Treatment Assay
Virus Pre-Treatment Assay
4.5.2. Antiproliferative Assays
Maintenance of Human Cell Cultures
Cytotoxicity Assay
4.5.3. MTT Assay
4.5.4. Biosurfactant Screening Assay by CTAB Agar Method
4.6. Chemical Profiling by Mass Spectrometry and Molecular Networking
4.7. Scale-Up Fermentation of I2R in SV2 SW Medium and Crude Extract Fractionation
4.8. Data-Dependent LC-HRMS/MS Analysis of the 90% MeOH Fraction
4.9. Methanolysis of the 90% MeOH Fraction and GC-MS Analysis of FAMEs
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naughton, L.M.; Romano, S.; O’Gara, F.; Dobson, A.D.W. Identification of Secondary Metabolite Gene Clusters in the Pseudovibrio Genus Reveals Encouraging Biosynthetic Potential toward the Production of Novel Bioactive Compounds. Front. Microbiol. 2017, 8, 1494. [Google Scholar] [CrossRef] [Green Version]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39 (Suppl. 2), W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gram, L. Silent clusters – speak up! Microb. Biotechnol. 2015, 8, 13–14. [Google Scholar] [CrossRef]
- van der Hooft, J.J.J.; Mohimani, H.; Bauermeister, A.; Dorrestein, P.C.; Duncan, K.R.; Medema, M.H. Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 2020, 49, 3297–3314. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Paulino, B.N.; Pessôa, M.G.; Mano, M.C.; Molina, G.; Neri-Numa, I.A.; Pastore, G.M. Current status in biotechnological production and applications of glycolipid biosurfactants. Appl. Microbiol. Biotechnol. 2016, 100, 10265–10293. [Google Scholar] [CrossRef]
- Jezierska, S.; Claus, S.; Van Bogaert, I. Yeast glycolipid biosurfactants. FEBS Lett. 2018, 592, 1312–1329. [Google Scholar] [CrossRef] [Green Version]
- Olasanmi, I.O.; Thring, R.W. The Role of Biosurfactants in the Continued Drive for Environmental Sustainability. Sustainability 2018, 10, 4817. [Google Scholar] [CrossRef] [Green Version]
- Hameş Tuna, E.E.; Fazilet, V.-S.; Kosaric, N. Patents on Biosurfactants and Future Trends. Biosurfactants Prod. Util.-Process. Technol. Econ. 2014, 159, 165. [Google Scholar] [CrossRef]
- Franzetti, A.; Gandolfi, I.; Bestetti, G.; Smyth, T.J.P.; Banat, I.M. Production and applications of trehalose lipid biosurfactants. Eur. J. Lipid Sci. Technol. 2010, 112, 617–627. [Google Scholar] [CrossRef]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannoni, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol 2010, 87, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Beaz-Hidalgo, R.; Hossain, M.J.; Liles, M.R. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2014, 2, e00927-14. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Comparative Genomics of the Rhodococcus Genus Shows Wide Distribution of Biodegradation Traits. Microorganisms 2020, 8, 774. [Google Scholar] [CrossRef]
- Peng, T.; Kan, J.; Hu, J.; Hu, Z. Genes and novel sRNAs involved in PAHs degradation in marine bacteria Rhodococcus sp. P14 revealed by the genome and transcriptome analysis. 3 Biotech 2020, 10, 140. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Sansegundo-Lobato, P.; Redondo-Nieto, M.; Suman, J.; Cajthaml, T.; Blanco-Romero, E.; Martin, M.; Uhlik, O.; Rivilla, R. Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp. WAY2 revealed by its complete genome sequence. Microb. Genom. 2020, 6, e000363. [Google Scholar] [CrossRef]
- Nachtigall, J.; Schneider, K.; Nicholson, G.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.-P. Two new aurachins from Rhodococcus sp. Acta 2259. J. Antibiot. 2010, 63, 567–569. [Google Scholar] [CrossRef]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a Cytotoxic Polyketide from a Streptomyces Strain Isolated from the Marine Sponge Halichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Awakawa, T.; Tuttle, R.N.; Letzel, A.C.; Kim, M.C.; Kudo, Y.; Fenical, W.; Moore, B.S.; Jensen, P.R. Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc. Natl. Acad. Sci. USA 2017, 114, E11121–E11130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelhardt, K.; Degnes, K.F.; Kemmler, M.; Bredholt, H.; Fjaervik, E.; Klinkenberg, G.; Sletta, H.; Ellingsen, T.E.; Zotchev, S.B. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 2010, 76, 4969–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zannella, C.; Giugliano, R.; Chianese, A.; Buonocore, C.; Vitale, G.A.; Sanna, G.; Sarno, F.; Manzin, A.; Nebbioso, A.; Termolino, P.; et al. Antiviral Activity of Vitis vinifera Leaf Extract against SARS-CoV-2 and HSV-1. Viruses 2021, 13, 1263. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, C.; Tedesco, P.; Vitale, G.A.; Palma Esposito, F.; Giugliano, R.; Monti, M.C.; D’Auria, M.V.; de Pascale, D. Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica). Mar. Drugs 2020, 18, 269. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Esch, S.W.; Morton, M.D.; Williams, T.D.; Buller, C.S. A novel trisaccharide glycolipid biosurfactant containing trehalose bears ester-linked hexanoate, succinate, and acyloxyacyl moieties: NMR and MS characterization of the underivatized structure. Carbohydr. Res. 1999, 319, 112–123. [Google Scholar] [CrossRef]
- Konishi, M.; Nishi, S.; Fukuoka, T.; Kitamoto, D.; Watsuji, T.-O.; Nagano, Y.; Yabuki, A.; Nakagawa, S.; Hatada, Y.; Horiuchi, J.-I. Deep-sea Rhodococcus sp. BS-15, Lacking the Phytopathogenic fas Genes, Produces a Novel Glucotriose Lipid Biosurfactant. Mar. Biotechnol. 2014, 16, 484–493. [Google Scholar] [CrossRef]
- Kügler, J.H.; Le Roes-Hill, M.; Syldatk, C.; Hausmann, R. Surfactants tailored by the class Actinobacteria. Front. Microbiol. 2015, 6, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demarque, D.P.; Crotti, A.E.M.; Vessecchi, R.; Lopes, J.L.C.; Lopes, N.P. Fragmentation reactions using electrospray ionization mass spectrometry: An important tool for the structural elucidation and characterization of synthetic and natural products. Nat. Prod. Rep. 2016, 33, 432–455. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xu, J.; Zhang, R.; Hao, Y.; He, J.; Chen, Y.; Jiao, G.; Abliz, Z. Strategy for Global Profiling and Identification of 2- and 3-Hydroxy Fatty Acids in Plasma by UPLC–MS/MS. Anal. Chem. 2020, 92, 5143–5151. [Google Scholar] [CrossRef] [PubMed]
- Fautz, E.; Rosenfelder, G.; Grotjahn, L. Iso-branched 2- and 3-hydroxy fatty acids as characteristic lipid constituents of some gliding bacteria. J. Bacteriol. 1979, 140, 852–858. [Google Scholar] [CrossRef] [Green Version]
- The Lipid Web. Available online: https://www.lipidmaps.org/resources/lipidweb (accessed on 16 June 2021).
- CHEMDATA.NIST.GOV. Available online: https://chemdata.nist.gov (accessed on 16 June 2021).
- Alvarez, H.M.; Luftmann, H.; Silva, R.A.; Cesari, A.C.; Viale, A.; Wältermann, M.; Steinbüchel, A. Identification of phenyldecanoic acid as a constituent of triacylglycerols and wax ester produced by Rhodococcus opacus PD630. Microbiology 2002, 148, 1407–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jae-Jun, S.; Mun-Hwan, C.; Sung-Chul, Y. Cometabolism of ω-Phenylalkanoic Acids with Butyric Acid for Efficient Production of Aromatic Polyesters in Pseudomonas putida BM01. J. Microbiol. Biotechnol. 2001, 11, 435–442. [Google Scholar]
- Tischler, D.; Niescher, S.; Kaschabek, S.R.; Schlömann, M. Trehalose phosphate synthases OtsA1 and OtsA2 of Rhodococcus opacus 1CP. FEMS Microbiol. Lett. 2013, 342, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Retamal-Morales, G.; Heine, T.; Tischler, J.S.; Erler, B.; Gröning, J.A.D.; Kaschabek, S.R.; Schlömann, M.; Levicán, G.; Tischler, D. Draft genome sequence of Rhodococcus erythropolis B7g, a biosurfactant producing actinobacterium. J. Biotechnol. 2018, 280, 38–41. [Google Scholar] [CrossRef]
- Delegan, Y.; Sargsyan, A.; Hovhannisyan, N.; Babayan, B.; Petrikov, K.; Vainstein, M. Analysis of genome sequence and trehalose lipid production peculiarities of the thermotolerant Gordonia strain. J. Basic Microbiol. 2020, 60, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Inaba, T.; Tokumoto, Y.; Miyazaki, Y.; Inoue, N.; Maseda, H.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Analysis of genes for succinoyl trehalose lipid production and increasing production in Rhodococcus sp. strain SD-74. Appl. Environ. Microbiol. 2013, 79, 7082–7090. [Google Scholar] [CrossRef] [Green Version]
- Palčeková, Z.; Angala, S.K.; Belardinelli, J.M.; Eskandarian, H.A.; Joe, M.; Brunton, R.; Rithner, C.; Jones, V.; Nigou, J.; Lowary, T.L.; et al. Disruption of the SucT acyltransferase in Mycobacterium smegmatis abrogates succinylation of cell envelope polysaccharides. J. Biol. Chem. 2019, 294, 10325–10335. [Google Scholar] [CrossRef]
- Schorn, M.A.; Alanjary, M.M.; Aguinaldo, K.; Korobeynikov, A.; Podell, S.; Patin, N.; Lincecum, T.; Jensen, P.R.; Ziemert, N.; Moore, B.S. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 2016, 162, 2075–2086. [Google Scholar] [CrossRef]
- Soldatou, S.; Eldjárn, G.H.; Ramsay, A.; van der Hooft, J.J.J.; Hughes, A.H.; Rogers, S.; Duncan, K.R. Comparative Metabologenomics Analysis of Polar Actinomycetes. Mar. Drugs 2021, 19, 103. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, J.; Hubmann, G.; Rosenthal, K.; Lütz, S. Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria. Biomolecules 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Vitale, G.A.; Sciarretta, M.; Cassiano, C.; Buonocore, C.; Festa, C.; Mazzella, V.; Núñez Pons, L.; D’Auria, M.V.; de Pascale, D. Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge. Int. J. Mol. Sci. 2020, 21, 6307. [Google Scholar] [CrossRef] [PubMed]
- Della Sala, G.; Teta, R.; Esposito, G.; Pawlik, J.R.; Mangoni, A.; Costantino, V. Zeamide, a Glycosylinositol Phosphorylceramide with the Novel Core Arap (1β→6) Ins Motif from the Marine Sponge Svenzea zeai. Molecules 2017, 22, 1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henkel, M.; Müller, M.M.; Kügler, J.H.; Lovaglio, R.B.; Contiero, J.; Syldatk, C.; Hausmann, R. Rhamnolipids as biosurfactants from renewable resources: Concepts for next-generation rhamnolipid production. Process Biochem. 2012, 47, 1207–1219. [Google Scholar] [CrossRef]
- Marchant, R.; Banat, I.M. Biosurfactants: A sustainable replacement for chemical surfactants? Biotechnol. Lett. 2012, 34, 1597–1605. [Google Scholar] [CrossRef]
- Tedesco, P.; Maida, I.; Palma Esposito, F.; Tortorella, E.; Subko, K.; Ezeofor, C.C.; Zhang, Y.; Tabudravu, J.; Jaspars, M.; Fani, R.; et al. Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica). Mar. Drugs 2016, 14, 83. [Google Scholar] [CrossRef] [Green Version]
- Chianese, G.; Esposito, F.P.; Parrot, D.; Ingham, C.; De Pascale, D.; Tasdemir, D. Linear Aminolipids with Moderate Antimicrobial Activity from the Antarctic Gram-Negative Bacterium Aequorivita sp. Mar. Drugs 2018, 16, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onwosi, C.O.; Aliyu, G.O.; Onu, C.J.; Chukwu, K.O.; Ndukwe, J.K.; Igbokwe, V.C. Microbial-derived glycolipids in the sustainable formulation of biomedical and personal care products: A consideration of the process economics towards commercialization. Process Biochem. 2021, 100, 124–139. [Google Scholar] [CrossRef]
- Tokumoto, Y.; Nomura, N.; Uchiyama, H.; Imura, T.; Morita, T.; Fukuoka, T.; Kitamoto, D. Structural characterization and surface-active properties of a succinoyl trehalose lipid produced by Rhodococcus sp. SD-74. J. Oleo Sci. 2009, 58, 97–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.N.R.L.; Farias, C.B.B.; Rufino, R.D.; Luna, J.M.; Sarubbo, L.A. Glycerol as substrate for the production of biosurfactant by Pseudomonas aeruginosa UCP0992. Colloids Surf. B Biointerfaces 2010, 79, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Putri, M.; Hertadi, R. Effect of Glycerol as Carbon Source for Biosurfactant Production by Halophilic Bacteria Pseudomonas Stutzeri BK-AB12. Procedia Chem. 2015, 16, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Ciapina, E.M.; Melo, W.C.; Santa Anna, L.M.; Santos, A.S.; Freire, D.M.; Pereira, N., Jr. Biosurfactant production by Rhodococcus erythropolis grown on glycerol as sole carbon source. Appl. Biochem. Biotechnol. 2006, 131, 880–886. [Google Scholar] [CrossRef]
- Giri, S.S.; Kim, H.J.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Park, S.C. Immunomodulatory Role of Microbial Surfactants, with Special Emphasis on Fish. Int. J. Mol. Sci. 2020, 21, 7004. [Google Scholar] [CrossRef]
- Christova, N.; Lang, S.; Wray, V.; Kaloyanov, K.; Konstantinov, S.; Stoineva, I. Production, Structural Elucidation, and In Vitro Antitumor Activity of Trehalose Lipid Biosurfactant from Nocardia farcinica Strain. J. Microbiol. Biotechnol. 2015, 25, 439–447. [Google Scholar] [CrossRef] [Green Version]
- Vollenbroich, D.; Ozel, M.; Vater, J.; Kamp, R.M.; Pauli, G. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis. Biol. J. Int. Assoc. Biol. Stand. 1997, 25, 289–297. [Google Scholar] [CrossRef]
- Jung, M.; Lee, S.; Kim, H. Recent studies on natural products as anti-HIV agents. Curr. Med. Chem. 2000, 7, 649–661. [Google Scholar] [CrossRef]
- Seydlová, G.; Svobodová, J. Review of surfactin chemical properties and the potential biomedical applications. Cent. Eur. J. Med. 2008, 3, 123–133. [Google Scholar] [CrossRef]
- Lang, S.; Wagner, F. Biological activities of biosurfactants. In Biosurfactants: Production-Properties-Applications; Kosaric, N., Ed.; CRC Press: Boca Raton, FL, USA, 1993; Volume 48, pp. 251–268. [Google Scholar]
- Uchida, Y.; Tsuchiya, R.; Chino, M.; Hirano, J.; Tabuchi, T.J.A. Extracellular Accumulation of Mono- and Di-Succinoyl Trehalose Lipids by a Strain of Rhodococcus erythropolis Grown on n-Alkanes. Agric. Biol. Chem. 1989, 53, 757–763. [Google Scholar] [CrossRef]
- Uchida, Y.; Misawa, S.; Nakahara, T.; Tabuchi, T. Factors Affecting the Production of Succinoyl Trehalose Lipids by Rhodococcus erythropolis SD-74 Grown on n-Alkanes. Agric. Biol. Chem. 1989, 53, 765–769. [Google Scholar] [CrossRef]
- Hoq, M.M.; Suzutani, T.; Toyoda, T.; Horiike, G.; Yoshida, I.; Azuma, M. Role of gamma delta TCR+ lymphocytes in the augmented resistance of trehalose 6,6′-dimycolate-treated mice to influenza virus infection. J. Gen. Virol. 1997, 78, 1597–1603. [Google Scholar] [CrossRef]
- Azuma, M.; Suzutani, T.; Sazaki, K.; Yoshida, I.; Sakuma, T.; Yoshida, T. Role of interferon in the augmented resistance of trehalose-6,6’-dimycolate-treated mice to influenza virus infection. J. Gen. Virol. 1987, 68, 835–843. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 September 2020).
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef]
- Singh, M.; Zannella, C.; Folliero, V.; Di Girolamo, R.; Bajardi, F.; Chianese, A.; Altucci, L.; Damasco, A.; Del Sorbo, M.R.; Imperatore, C.; et al. Combating Actions of Green 2D-Materials on Gram Positive and Negative Bacteria and Enveloped Viruses. Front. Bioeng. Biotechnol. 2020, 8, 569967. [Google Scholar] [CrossRef]
- Romero, J.R.; Gross, T.; Abromowitch, M.; Jung, L. Pleconaril Treatment of Vaccine-Acquired Poliovirus. Pediatr. Res. 1999, 45, 173. [Google Scholar] [CrossRef] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef]
- Della Sala, G.; Mangoni, A.; Costantino, V.; Teta, R. Identification of the Biosynthetic Gene Cluster of Thermoactinoamides and Discovery of New Congeners by Integrated Genome Mining and MS-Based Molecular Networking. Front. Chem. 2020, 8, 397. [Google Scholar] [CrossRef]
- Johnson, M.B.; Wen, Z. Production of Biodiesel Fuel from the Microalga Schizochytrium limacinum by Direct Transesterification of Algal Biomass. Energy Fuels 2009, 23, 5179–5183. [Google Scholar] [CrossRef]


| Attribute | Value |
|---|---|
| Genome size (bp) | 5,290,284 |
| DNA G+C (%) | 64.01 |
| Number of contigs | 72 |
| Longest contig length (bp) | 336,301 |
| Shortest contig length (bp) | 1117 |
| Average contig length (bp) | 73,476 |
| N50 (bp) | 195,979 |
| Number of Coding Sequences | 5210 |
| RNA genes (tRNA + rRNA) | 52 |
| Type | Contig | Location (nt) | Most Similar Known Cluster | Similarity (%) |
|---|---|---|---|---|
| Saccharide—NRPS | 1 | 29,248–91,486 | Coelichelin | 27 |
| Saccharide | 1 | 313,381–336,301 | Macrotetrolide | 33 |
| T2PKS—saccharide | 2 | 47,197–119,676 | Mayamycin | 63 |
| Saccharide | 2 | 236,836–273,343 | TP-1161 | 8 |
| NRPS—redox-cofactor—saccharide | 3 | 1–92,466 | – | – |
| Ectoine | 4 | 116,604–127,002 | Ectoine | 50 |
| Saccharide—T1PKS | 5 | 1–59,796 | Selvamicin | 11 |
| Fatty acid | 5 | 148,163–167,687 | – | – |
| Fatty acid | 5 | 212,303–233,406 | – | – |
| Saccharide | 6 | 107,905–134,028 | – | – |
| Saccharide—terpene | 6 | 144,642–204,805 | Isorenieratene | 42 |
| Saccharide | 7 | 153–31,205 | – | – |
| Saccharide | 7 | 42,961–63,566 | Tetronasin | 3 |
| Saccharide | 7 | 96,022–137,264 | Rimosamide | 14 |
| NRPS-like | 7 | 142,446–185,022 | – | – |
| Terpene | 8 | 92,586–113,764 | SF2575 | 6 |
| Saccharide | 9 | 108,630–145,274 | ECO-02301 | 7 |
| Saccharide | 10 | 73,245–94,928 | Streptovaricin | 4 |
| Saccharide | 10 | 98,951–137,834 | – | – |
| PKS-like—amglyccycl * | 12 | 158,941–192,419 | – | – |
| Furan—fatty acid | 13 | 13,932–35,940 | Diisonitrile antibiotic SF2768 | 11 |
| NAPAA * | 15 | 1–30,389 | – | – |
| hglE-KS * | 16 | 63,473–105,151 | Vazabitide A | 10 |
| NRPS | 17 | 1–29,241 | Atratumycin | 5 |
| NRPS—saccharide | 18 | 1–79,116 | – | – |
| Butyrolactone | 21 | 68,428–79,324 | – | – |
| Arylpolyene—T1PKS—fatty acid—PKS-like | 24 | 1–67,243 | Abyssomicin C/Atrop-Abyssomicin C | 7 |
| NRPS—saccharide | 26 | 1–59,270 | Heterobactin A/Heterobactin S2 | 63 |
| NRPS | 27 | 1–54,662 | Siamycin I | 8 |
| Fatty acid—saccharide | 28 | 3562–54,528 | Bottromycin A2 | 9 |
| NRPS-like | 29 | 27,485–54,026 | – | – |
| Saccharide | 32 | 1–17,712 | – | – |
| Saccharide | 32 | 34,798–52,216 | Tetrocarcin A | 4 |
| NRPS | 39 | 1–22,870 | – | – |
| NRPS | 43 | 1–12,753 | Atratumycin | 5 |
| Saccharide | 54 | 1–5008 | – | – |
| Rt (min.) | [M − H]– | m/z | Primary Acyl Chains | Secondary Acyl Chains a | |||||
|---|---|---|---|---|---|---|---|---|---|
| disaccharide succinic diesters | 14.6 | C26H43O16 | 611.2546 | 3-OH-C10 | - | ||||
| 15.4 | C25H41O15 | 581.2424 | C9 | - | |||||
| 16 | C27H45O16 | 625.2685 | 3-OH-C11 | - | |||||
| 16.3 | C26H41O15 | 593.2427 | C10:1 | - | |||||
| 16.7 | C26H43O15 | 595.2585 | C10 | - | |||||
| 18 | C32H45O17 | 701.2633 | 3-OH-C8 b | PhAc | |||||
| 19.3 | C30H51O16 | 667.3155 | 3-OH-C14 | - | |||||
| 19.9 | C34H49O17 | 729.2948 | 3-OH-C10 | PhAc | |||||
| disaccharide succinic triesters | 16.6 | C34H49O18 | 745.2901 | 3-OH-C8 | 3-OH-PhBu | - | |||
| 16.9 | C32H45O17 | 701.2634 | 3-OH-C8 | PhAc | - | ||||
| 17.5 | C32H53O18 | 725.3209 | 3-OH-C8 | 3-OH-C8 | - | ||||
| 17.8 | C40H59O20 | 859.3571 | 3-OH-C8 | diOH-C8 | PhAc | ||||
| 18.5 | C33H55O18 | 739.3367 | 3-OH-C9 | 3-OH-C8 | - | ||||
| 19 | C34H49O17 | 729.2944 | 3-OH-C10 | PhAc | - | ||||
| 19.4 | C34H57O18 | 753.3523 | 3-OH-C10 | 3-OH-C8 | - | ||||
| 19.8 | C38H55O19 | 815.3307 | 3-OH-C8 | 3-OH-C6 | PhAc c | ||||
| 20.8 | C39H57O19 | 829.3463 | 3-OH-C8 | 3-OH-C7 | PhAc c | ||||
| 20.9 | C42H55O19 | 863.3309 | 3-OH-C8 | 3-OH-PhBu | PhAc | ||||
| 21.6 | C40H59O19 | 843.3619 | 3-OH-C8 | 3-OH-C8 | PhAc | ||||
| 22.4 | C41H61O19 | 857.3775 | 3-OH-C9 | 3-OH-C8 | PhAc c | ||||
| 25.2 | C44H67O19 | 899.4243 | 3-OH-C10 | 3-OH-C10 | PhAc | ||||
| trisaccharide succinic diesters | 17.2 | C38H55O22 | 863.3155 | 3-OH-C8 | PhAc | ||||
| trisaccharide succinic triesters | 18 | C40H67O23 | 915.4041 | 3-OH-C10 | 3-OH-C8 | - | |||
| 18.4 | C48H73O25 | 1049.4406 | 3-OH-C10 | diOH-C8 | PhAc | ||||
| 19.5 | C48H65O24 | 1025.3827 | 3-OH-C8 | 3-OH-PhBu | PhAc | ||||
| 20.1 | C44H73O24 | 985.4456 | 3-OH-C8 | 3-OH-C8 | C6 | ||||
| 20.1 | C46H69O24 | 1005.4138 | 3-OH-C8 | 3-OH-C8 | PhAc | ||||
| 20.8 | C47H71O24 | 1019.4294 | 3-OH-C9 | 3-OH-C8 | PhAc c | ||||
| 21.5 | C48H73O24 | 1033.4447 | 3-OH-C10 | 3-OH-C8 | PhAc c | ||||
| 22.2 | C49H75O24 | 1047.4602 | 3-OH-C10 | 3-OH-C9 | PhAc | ||||
| 22.2 | C49H75O24 | 1047.4602 | 3-OH-C11 | 3-OH-C8 | PhAc | ||||
| 23.1 | C50H77O24 | 1061.4761 | 3-OH-C10 | 3-OH-C10 | PhAc | ||||
| 23.6 | C55H77O25 | 1137.4707 | 3-OH-C9 | 3-OH-C8 | PhAc | PhAc d | |||
| 23.7 | C52H79O25 | 1103.4865 | 3-OH-C8 | 3-OH-C8 | PhAc | C6 d | |||
| 24.6 | C53H81O25 | 1117.5018 | 3-OH-C9 | 3-OH-C8 | PhAc | C6 d | |||
| 26.3 | C55H85O25 | 1145.53296 | 3-OH-C11 | 3-OH-C8 | PhAc | C6 d | |||
| 26.3 | C55H85O25 | 1145.53296 | 3-OH-C10 | 3-OH-C9 | PhAc | C6 d | |||
| trisaccharide succinic tetraesters | 27.9 | C54H83O24 | 1115.5221 | 3-OH-C8 | C10 | C6 e | PhAc | ||
| 29.4 | C58H89O26 | 1201.5579 | 3-OH-C8 | 3-OH-C8 | C6 e | PhAc | C6 d | ||
| 3-Hydroxy FAMEs | Relative Abundance (%) |
|---|---|
| 3-OH-C6 | 0.2 |
| 3-OH-C7, branched | 0.1 |
| 3-OH-C7 | 0.5 |
| 3-OH-C8, branched | 6.0 |
| 3-OH-C8 | 31.7 |
| 3-OH-C9, branched | 0.7 |
| 3-OH-C9 | 8.0 |
| 3-OH-C10, branched | 3.0 |
| 3-OH-C10 | 36.1 |
| 3-OH-4-phenylbutanoate | 2.2 |
| 3-OH-C11 | 2.8 |
| 3-OH-C12 | 7.6 |
| 3-OH-C13 | 0.2 |
| 3-OH-C14 | 1.0 |
| Contig | Start | Strand | Length | Putative Gene | Function |
|---|---|---|---|---|---|
| contig4 | 178,279 | − | 2043 | − | Trehalase |
| contig6 | 117,624 | + | 2274 | treX | Glycogen debranching enzyme |
| contig6 | 119,901 | + | 2418 | treY | Putative maltooligosyl trehalose synthase |
| contig6 | 122,315 | + | 1740 | treZ | Malto-oligosyltrehalose trehalohydrolase |
| contig15 | 69,438 | − | 2550 | otsB | Trehalose-6-phosphate phosphatase |
| contig18 | 99,608 | − | 2181 | treS | Trehalose synthase |
| contig19 | 37,715 | + | 3192 | otsB | Trehalose-6-phosphate phosphatase |
| contig23 | 53,441 | + | 1458 | otsA | Trehalose-6-phosphate synthase |
| contig44 | 6907 | + | 1068 | fbaA | Fructose-bisphosphate aldolase |
| contig44 | 9581 | + | 1041 | fbaA | Fructose-bisphosphate aldolase |
| contig6 | 115,388 | + | 2202 | sucT | Putative succinoyl transferase |
| contig5 | 46,744 | − | 1887 | − | Trehalose O-mycolyltransferase |
| contig9 | 137,187 | + | 1461 | papA3 | Acyltransferase papA3 |
| contig9 | 138,665 | + | 1431 | papA1 | SL659 acyltransferase papA1 |
| contig9 | 140,099 | + | 1404 | papA3 | Acyltransferase papA3 |
| contig9 | 143,010 | + | 1488 | papA3 | Acyltransferase papA3 |
| contig2 | 114,384 | + | 1071 | paaK | Phenylacetate-CoA oxygenase/reductase, PaaK subunit |
| contig3 | 53,842 | + | 1044 | paaK | Phenylacetate-CoA oxygenase/reductase, PaaK subunit |
| contig3 | 219,048 | + | 1449 | feaB | Phenylacetaldehyde dehydrogenase |
| contig4 | 26,516 | − | 2238 | katG | Catalase-peroxidase 2 |
| contig4 | 233,312 | − | 204 | paaK | Phenylacetate-CoA oxygenase/reductase, PaaK subunit |
| contig14 | 61,296 | + | 795 | amiE | Aliphatic amidase AmiE |
| contig20 | 84,586 | − | 840 | amiE | Aliphatic amidase AmiE |
| contig7 | 14,325 | + | 3195 | mmpL3 | Trehalose monomycolate exporter MmpL3 |
| contig10 | 71,540 | − | 1371 | lpqY | Trehalose-binding lipoprotein LpqY |
| contig12 | 94,464 | + | 2226 | mmpL3 | Trehalose monomycolate exporter MmpL3 |
| contig17 | 98,444 | − | 2136 | mmpL3 | Trehalose monomycolate exporter MmpL3 |
| contig5 | 96,103 | + | 1053 | sugC | Trehalose import ATP-binding protein SugC |
| contig5 | 265,744 | + | 1050 | sugA | Trehalose transport system permease protein SugA |
| contig5 | 266,790 | + | 822 | sugB | Trehalose transport system permease protein SugB |
| contig7 | 42,107 | − | 1101 | sugC | Trehalose import ATP-binding protein SugC |
| contig7 | 42,961 | − | 840 | sugB | Trehalose transport system permease protein SugB |
| contig7 | 43,911 | − | 951 | sugA | Trehalose transport system permease protein SugA |
| contig7 | 56,201 | + | 948 | sugA | Trehalose transport system permease protein SugA |
| contig7 | 100,213 | − | 1083 | sugC | Trehalose import ATP-binding protein SugC |
| contig10 | 68,393 | − | 1170 | sugC | Trehalose import ATP-binding protein SugC |
| contig10 | 69,232 | − | 834 | sugB | Trehalose transport system permease protein SugB |
| contig10 | 70,173 | − | 942 | sugA | Trehalose transport system permease protein SugA |
| contig13 | 120,539 | − | 1077 | sugC | Trehalose import ATP-binding protein SugC |
| contig20 | 57,846 | − | 1149 | sugC | Trehalose import ATP-binding protein SugC |
| contig20 | 59,851 | − | 867 | sugB | Trehalose transport system permease protein SugB |
| contig20 | 60,832 | − | 978 | sugA | Trehalose transport system permease protein SugA |
| Virus | Family | Nucleic Acid | Symmetry | Envelope | Dimensions |
|---|---|---|---|---|---|
| HSV-1/GFP | Herpesviridae | dsDNA | icosahedral | yes | 155–240 nm |
| HCoV-229E | Coronaviridae | ssRNA(+) | helical | yes | 80–120 nm |
| HCoV-OC43 | Coronoviridae | ssRNA(+) | helical | yes | 80–120 nm |
| PV-1 | Picornaviridae | ssRNA(+) | icosahedral | no | 30 nm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma Esposito, F.; Giugliano, R.; Della Sala, G.; Vitale, G.A.; Buonocore, C.; Ausuri, J.; Galasso, C.; Coppola, D.; Franci, G.; Galdiero, M.; et al. Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus. Int. J. Mol. Sci. 2021, 22, 9055. https://doi.org/10.3390/ijms22169055
Palma Esposito F, Giugliano R, Della Sala G, Vitale GA, Buonocore C, Ausuri J, Galasso C, Coppola D, Franci G, Galdiero M, et al. Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus. International Journal of Molecular Sciences. 2021; 22(16):9055. https://doi.org/10.3390/ijms22169055
Chicago/Turabian StylePalma Esposito, Fortunato, Rosa Giugliano, Gerardo Della Sala, Giovanni Andrea Vitale, Carmine Buonocore, Janardhan Ausuri, Christian Galasso, Daniela Coppola, Gianluigi Franci, Massimiliano Galdiero, and et al. 2021. "Combining OSMAC Approach and Untargeted Metabolomics for the Identification of New Glycolipids with Potent Antiviral Activity Produced by a Marine Rhodococcus" International Journal of Molecular Sciences 22, no. 16: 9055. https://doi.org/10.3390/ijms22169055







