Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications
Abstract
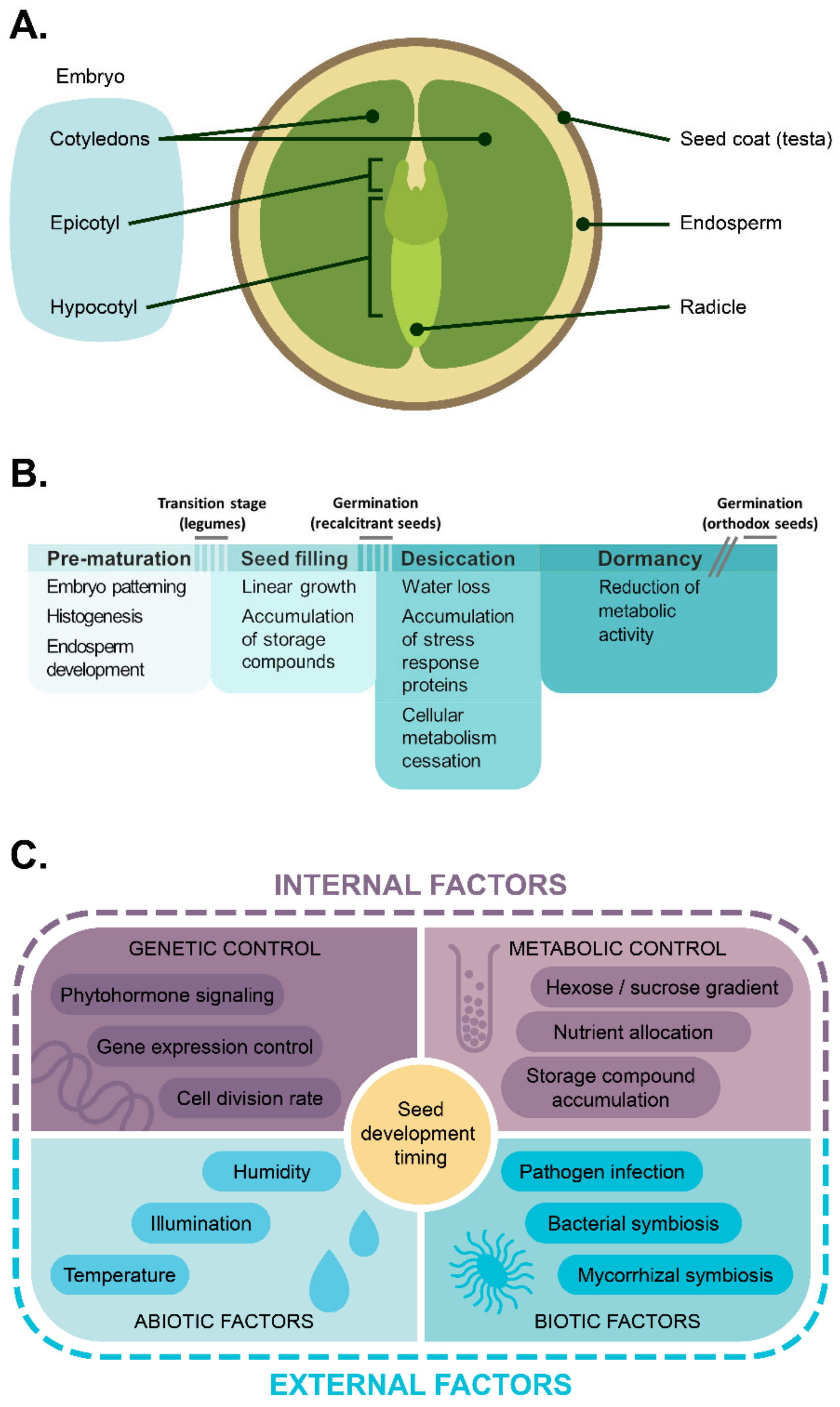
1. Introduction
2. Cell Proliferation during Embryogenesis
3. Endoreduplication and Cell Expansion
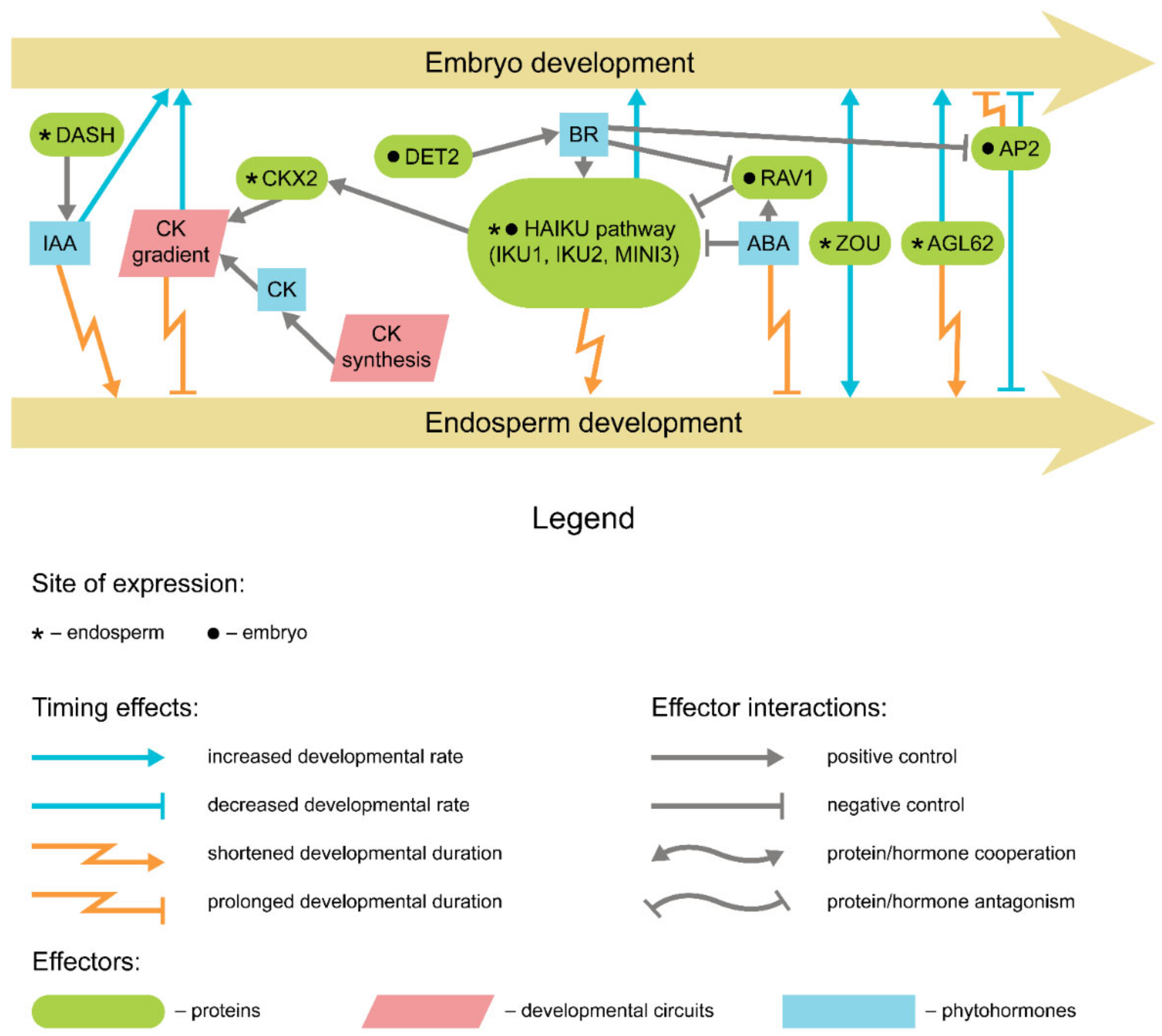
4. Genetic Control of Seed Maturation
5. Endosperm and Seed Coat Development
6. Two-Membrane Organelle Functioning and Energy Metabolism
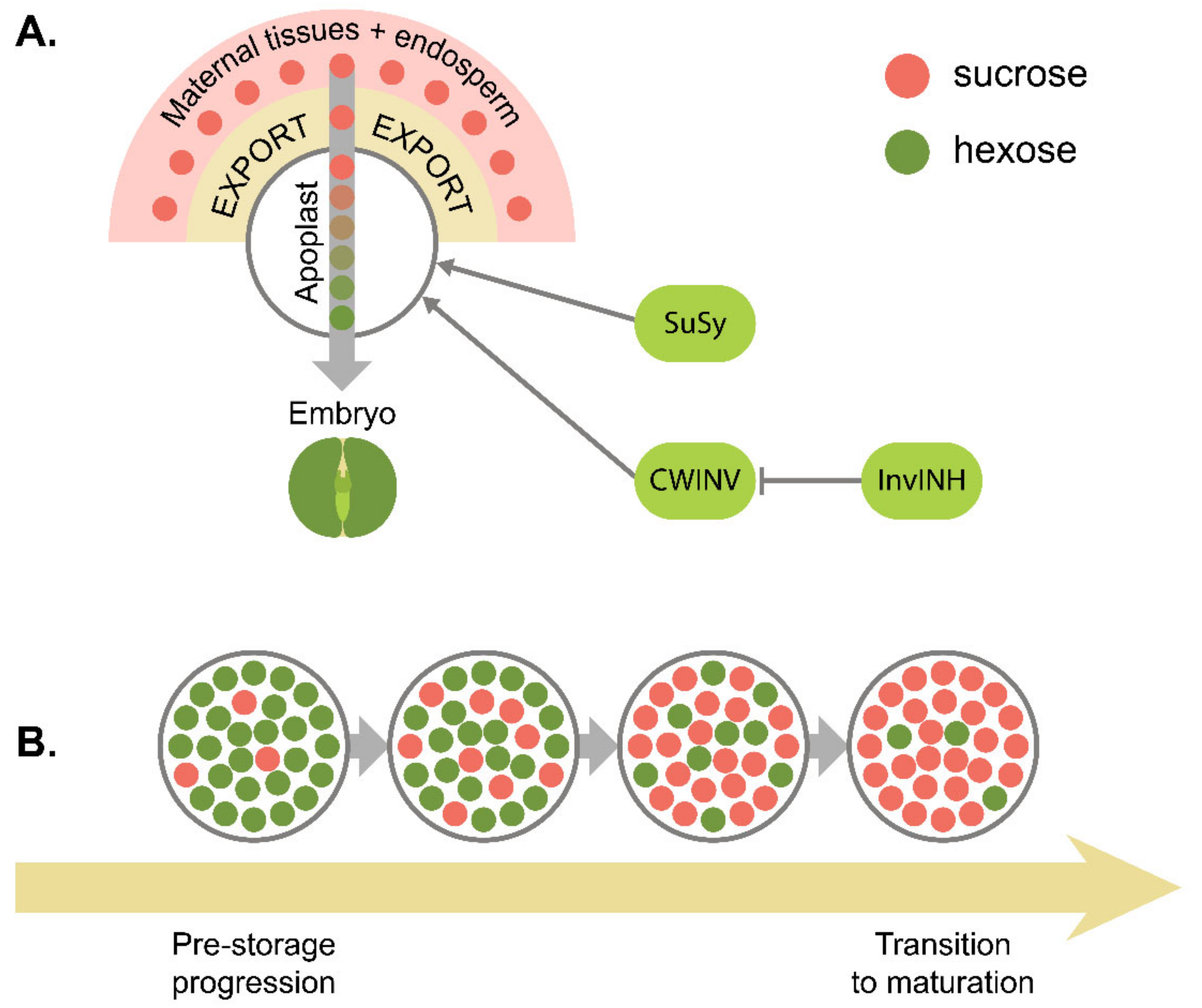
7. Metabolic Control of Seed Development
8. Environmental Factors Affecting Seed Development Rate
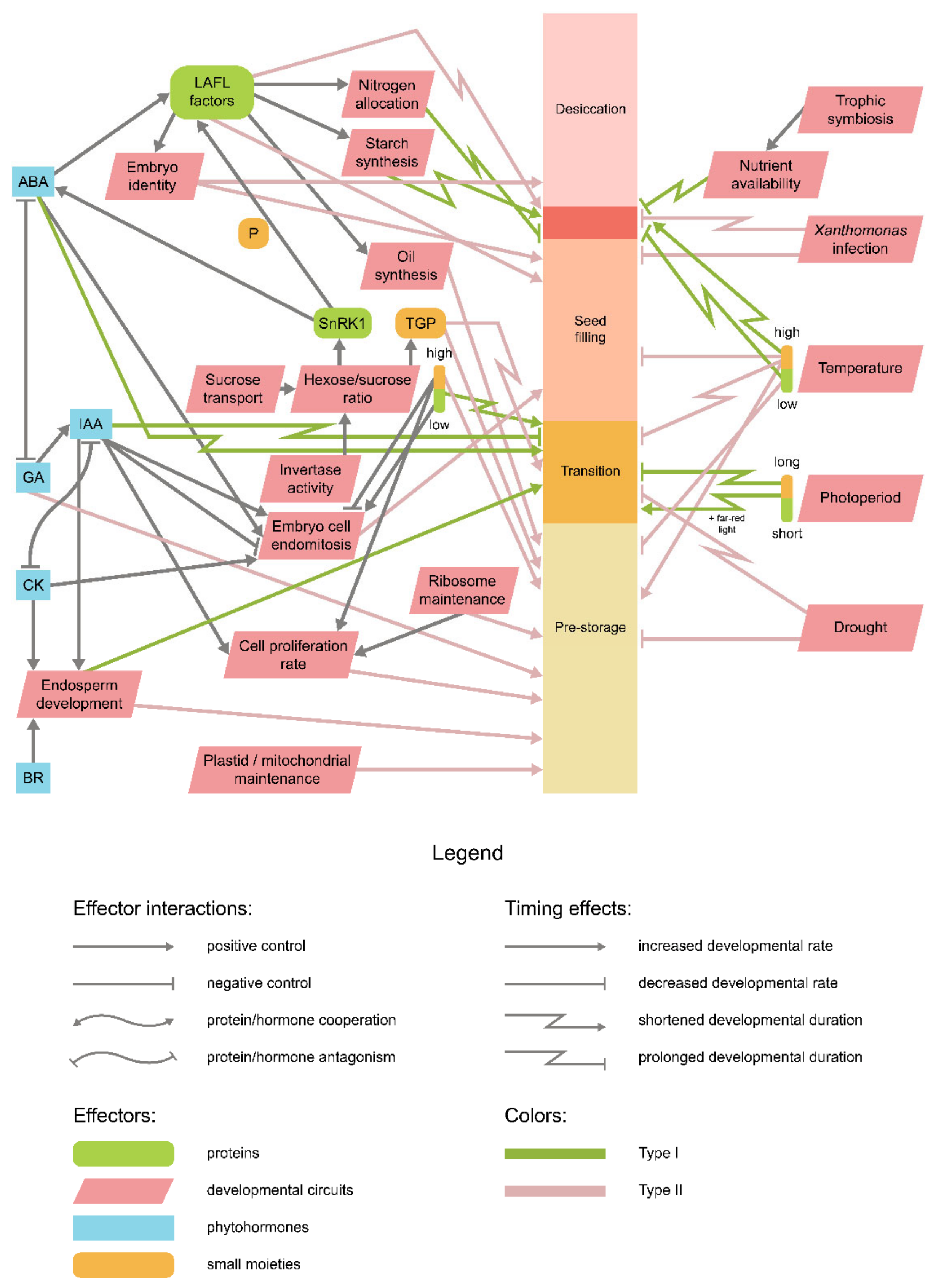
9. Is There an Integrative Scheme of Seed Development Timing Control?
10. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Le, B.H.; Wagmaister, J.A.; Kawashima, T.; Bui, A.Q.; Harada, J.J.; Goldberg, R.B. Using genomics to study legume seed development. Plant Physiol. 2007, 144, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Capron, A.; Chatfield, S.; Provart, N.; Berleth, T. Embryogenesis: Pattern formation from a single cell. Arab. Book 2009, 7, e0126. [Google Scholar] [CrossRef]
- Olsen, O.-A. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 2004, 16, S214–S227. [Google Scholar] [CrossRef]
- Raz, V.; Bergervoet, J.; Koornneef, M. Sequential steps for developmental arrest in Arabidopsis seeds. Development 2001, 128, 243–252. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2016, 68, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Farrant, J.; Pammenter, N.W.; Berjak, P. Seed development in relation to desiccation tolerance: A comparison between desiccation-sensitive (recalcitrant) seeds of Avicennia marina and desiccation-tolerant types. Seed Sci. Res. 1993, 3, 1–13. [Google Scholar] [CrossRef]
- Farnsworth, E.J.; Farrant, J. Reductions in abscisic acid are linked with viviparous reproduction in mangroves. Am. J. Bot. 1998, 85, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Berjak, P.; Pammenter, N. Seed recalcitrance—Current perspectives. South Afr. J. Bot. 2001, 67, 79–89. [Google Scholar] [CrossRef]
- Nikolov, L.A.; Tomlinson, P.B.; Manickam, S.; Endress, P.K.; Kramer, E.M.; Davis, C.C. Holoparasitic Rafflesiaceae possess the most reduced endophytes and yet give rise to the world’s largest flowers. Ann. Bot. 2014, 114, 233–242. [Google Scholar] [CrossRef]
- Mitchell, J.; Johnston, I.G.; Bassel, G.W. Variability in seeds: Biological, ecological, and agricultural implications. J. Exp. Bot. 2016, 68, 809–817. [Google Scholar] [CrossRef]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant Sci. 2014, 5, 412. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Rodríguez-Gacio, M.D.C.; Matilla-Vázquez, M.A.; Matilla, A.J. Seed dormancy and ABA signaling. Plant Signal. Behav. 2009, 4, 1035–1048. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Robert, H.S.; Park, C.; Gutièrrez, C.L.; Wójcikowska, B.; Pěnčík, A.; Novák, O.; Chen, J.; Grunewald, W.; Dresselhaus, T.; Friml, J.; et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 2018, 4, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Van Wuytswinkel, O.; Castelain, M.; Bellini, C. Combined networks regulating seed maturation. Trends Plant Sci. 2007, 12, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Perry, S.E. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013, 161, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Devic, M.; Roscoe, T. Seed maturation: Simplification of control networks in plants. Plant Sci. 2016, 252, 335–346. [Google Scholar] [CrossRef]
- Han, J.-D.; Li, X.; Jiang, C.-K.; Wong, G.K.-S.; Rothfels, C.J.; Rao, G.-Y. Evolutionary analysis of the LAFL genes involved in the land plant seed maturation program. Front. Plant Sci. 2017, 8, 439. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Devic, M.; Roscoe, T.J.; Bouyer, D.; Zhou, D.-X.; Boulard, C.; Baud, S.; Dubreucq, B. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 2018, 31, 291–307. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Ingouff, M.; Haseloff, J.; Berger, F. Polycomb group genes control developmental timing of endosperm. Plant J. 2005, 42, 663–674. [Google Scholar] [CrossRef]
- Nodine, M.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P.D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- An, Y.-Q.C.; Goettel, W.; Han, Q.; Bartels, A.; Liu, Z.; Xiao, W. Dynamic changes of genome-wide DNA methylation during soybean seed development. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Wobus, U.; Weber, H. Sugars as signal molecules in plant seed development. Biol. Chem. 1999, 380, 937–944. [Google Scholar] [CrossRef]
- Borisjuk, L.; Rolletschek, H.; Wobus, U.; Weber, H. Differentiation of legume cotyledons as related to metabolic gradients and assimilate transport into seeds. J. Exp. Bot. 2003, 54, 503–512. [Google Scholar] [CrossRef]
- Gomez, L.D.; Baud, S.; Gilday, A.; Li, Y.; Graham, I.A. Delayed embryo development in the Arabidopsis trehalose-6-phosphate synthase1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 2006, 46, 69–84. [Google Scholar] [CrossRef]
- Baud, S.; Wuillème, S.; Dubreucq, B.; De Almeida, A.; Vuagnat, C.; Lepiniec, L.; Miquel, M.; Rochat, C. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 2007, 52, 405–419. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoët, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef]
- Bueckert, R.A.; Wagenhoffer, S.; Hnatowich, G.; Warkentin, T. Effect of heat and precipitation on pea yield and reproductive performance in the field. Can. J. Plant Sci. 2015, 95, 629–639. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Hanumantharao, B.; Nair, R.M.; Prasad, P.V.V.; Kumar, S.; Gaur, P.; Farooq, M.; Siddique, K.; Varshney, R.; et al. Food legumes and rising temperatures: Effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef] [PubMed]
- Larmure, A.; Munier-Jolain, N.G. High temperatures during the seed-filling period decrease seed nitrogen amount in pea (Pisum sativum L.): Evidence for a sink limitation. Front. Plant Sci. 2019, 10, 1608. [Google Scholar] [CrossRef] [PubMed]
- Ribalta, F.M.; Pazos-Navarro, M.; Edwards, K.; Ross, J.J.; Croser, J.; Ochatt, S.J. Expression patterns of key hormones related to pea (Pisum sativum L.) embryo physiological maturity shift in response to accelerated growth conditions. Front. Plant Sci. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Terrasson, E.; Darrasse, A.; Righetti, K.; Buitink, J.; Lalanne, D.; Vu, B.L.; Pelletier, S.; Bolingue, W.; Jacques, M.-A.; Leprince, O. Identification of a molecular dialogue between developing seeds of Medicago truncatula and seedborne xanthomonads. J. Exp. Bot. 2015, 66, 3737–3752. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Q.; Cregan, P.B.; Nelson, R.L.; Wang, X.; Wu, J.; Jiang, G.-L. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef]
- Copley, T.R.; Duceppe, M.-O.; O’Donoughue, L.S. Identification of novel loci associated with maturity and yield traits in early maturity soybean plant introduction lines. BMC Genom. 2018, 19, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Li, Y.-Q.; Wu, H.-Y.; Hu, B.; Zheng, J.-J.; Zhai, H.; Lv, S.-X.; Liu, X.-L.; Chen, X.; Qiu, H.-M.; et al. Genotyping of soybean cultivars with medium-density array reveals the population structure and QTNs underlying maturity and seed traits. Front. Plant Sci. 2018, 9, 610. [Google Scholar] [CrossRef]
- Kamfwa, K.; Cichy, K.; Kelly, J.D. Genome-wide association study of agronomic traits in common bean. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Li, Y.; Yang, K.; Yang, W.; Chu, L.; Chen, C.; Zhao, B.; Li, Y.; Jian, J.; Yin, Z.; Wang, T.; et al. Identification of QTL and qualitative trait loci for agronomic traits using SNP markers in the Adzuki bean. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Gali, K.K.; Sackville, A.; Tafesse, E.G.; Lachagari, V.R.; McPhee, K.; Hybl, M.; Mikić, A.; Smykal, P.; McGee, R.; Burstin, J.; et al. Genome-wide association mapping for agronomic and seed quality traits of field pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 1538. [Google Scholar] [CrossRef]
- Ribalta, F.M.; Pazos-Navarro, M.; Nelson, K.; Edwards, K.; Ross, J.J.; Bennett, R.; Munday, C.; Erskine, W.; Ochatt, S.J.; Croser, J. Precocious floral initiation and identification of exact timing of embryo physiological maturity facilitate germination of immature seeds to truncate the lifecycle of pea. Plant Growth Regul. 2016, 81, 345–353. [Google Scholar] [CrossRef]
- Croser, J.; Ribalta, F.; Navarro, M.P.; Munday, C.; Bennett, R.; Kaur, P.; Ochatt, S. In vitro-assisted compression of breeding cycles. In Biotechnologies of Crop Improvement; Gosal, S.S., Hussain, W.S., Eds.; Springer: Cham, Switzerland, 2018; Volume 1, pp. 463–486. [Google Scholar]
- Möller, B.; Weijers, D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a001545. [Google Scholar] [CrossRef]
- Bayer, M.; Slane, D.; Jürgens, G. Early plant embryogenesis—Dark ages or dark matter? Curr. Opin. Plant Biol. 2017, 35, 30–36. [Google Scholar] [CrossRef]
- Wang, K.; Chen, H.; Miao, Y.; Bayer, M. Square one: Zygote polarity and early embryogenesis in flowering plants. Curr. Opin. Plant Biol. 2019, 53, 128–133. [Google Scholar] [CrossRef]
- Munier-Jolain, N.G.; Ney, B. Seed growth rate in grain legumes. Seed growth rate depends on cotyledon cell number. J. Exp. Bot. 1998, 49, 1971–1976. [Google Scholar] [CrossRef]
- Powell, A.E.; Lenhard, M. Control of organ size in plants. Curr. Biol. 2012, 22, R360–R367. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Borisjuk, L.; Wobus, U. Controlling seed development and seed size in Vicia faba: A role for seed coat-associated invertases and carbohydrate state. Plant J. 1996, 10, 823–834. [Google Scholar] [CrossRef]
- Bandyopadhyay, K.; Ulucay, O.; Şakiroğlu, M.; Udvardi, M.K.; Verdier, J. Analysis of large seeds from three different Medicago truncatula ecotypes reveals a potential role of hormonal balance in final size determination of legume grains. Int. J. Mol. Sci. 2016, 17, 1472. [Google Scholar] [CrossRef] [PubMed]
- Lemontey, C.; Mousset-Déclas, C.; Munier-Jolain, N.; Boutin, J. Maternal genotype influences pea seed size by controlling both mitotic activity during early embryogenesis and final endoreduplication level/cotyledon cell size in mature seed. J. Exp. Bot. 2000, 51, 167–175. [Google Scholar] [CrossRef]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A Unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of short hypocotyl under BLUE1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Valkai, I.; Labhane, N.M.; Koczka, L.; Andrási, N.; Klement, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; et al. CRK5 protein kinase contributes to the progression of embryogenesis of Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6120. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.D.L.P.; Costas, C.; Sequeira-Mendes, J.; Gutiérrez, C. Regulating DNA replication in plants. Cold Spring Harb. Perspect. Biol. 2012, 4, a010140. [Google Scholar] [CrossRef]
- Dante, R.A.; Larkins, B.A.; Sabelli, P.A. Cell cycle control and seed development. Front. Plant Sci. 2014, 5, 493. [Google Scholar] [CrossRef] [PubMed]
- Tank, J.G.; Pandya, R.V.; Thaker, V.S. Phytohormones in regulation of the cell division and endoreduplication process in the plant cell cycle. RSC Adv. 2014, 4, 12605. [Google Scholar] [CrossRef]
- Jenik, P.; Jurkuta, R.E.; Barton, M.K. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase ε. Plant Cell 2005, 17, 3362–3377. [Google Scholar] [CrossRef]
- O’Neill, J.P.; Colon, K.T.; Jenik, P.D. The onset of embryo maturation in Arabidopsis is determined by its developmental stage and does not depend on endosperm cellularization. Plant J. 2019, 99, 286–301. [Google Scholar] [CrossRef]
- Collins, C.; Dewitte, W.; Murray, J.A.H. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 2012, 63, 3571–3586. [Google Scholar] [CrossRef]
- Dewitte, W.; Riou, C.; Scofield, S.; Healy, J.M.S.; Jacqmard, A.; Kilby, N.J.; Murray, J.A.H. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 2002, 15, 79–92. [Google Scholar] [CrossRef]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Isono, E. The COP9 signalosome and its role in plant development. Eur. J. Cell Biol. 2010, 89, 157–162. [Google Scholar] [CrossRef]
- Franciosini, A.; Moubayidin, L.; Du, K.; Matari, N.H.; Boccaccini, A.; Butera, S.; Vittorioso, P.; Sabatini, S.; Jenik, P.D.; Costantino, P.; et al. The COP9 signalosome is required for postembryonic meristem maintenance in Arabidopsis thaliana. Mol. Plant 2015, 8, 1623–1634. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Betsch, L.; Boltz, V.; Brioudes, F.; Pontier, G.; Girard, V.; Savarin, J.; Wipperman, B.; Chambrier, P.; Tissot, N.; Benhamed, M.; et al. TCTP and CSN4 control cell cycle progression and development by regulating CULLIN1 neddylation in plants and animals. PLoS Genet. 2019, 15, e1007899. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, Y.-L. Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Meitzel, T.; Radchuk, R.; Nunes-Nesi, A.; Fernie, A.R.; Link, W.; Weschke, W.; Weber, H. Hybrid embryos of Vicia faba develop enhanced sink strength, which is established during early development. Plant J. 2010, 65, 517–531. [Google Scholar] [CrossRef]
- Wang, H.-J.; Hsu, Y.-W.; Guo, C.-L.; Jane, W.-N.; Wang, H.; Jiang, L.; Jauh, G.-Y. VPS36-dependent multivesicular bodies are critical for plasmamembrane protein turnover and vacuolar biogenesis. Plant Physiol. 2016, 173, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The auxin response factor 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by auxin response factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef]
- Swain, S.; Ross, J.; Reid, J.; Kamiya, Y. Gibberellins and pea seed development. Planta 1995, 195, 426–433. [Google Scholar] [CrossRef]
- Swain, S.M.; Reid, J.B.; Kamiya, Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997, 12, 1329–1338. [Google Scholar] [CrossRef]
- Quesnelle, P.E.Q.E.; Emery, N. Cis-cytokinins that predominate in Pisum sativum during early embryogenesis will accelerate embryo growth in vitro. Can. J. Bot. 2007, 85, 91–103. [Google Scholar] [CrossRef]
- Levi, M.; Brusa, P.; Chiatante, D.; Sparvoli, E. Cell cycle reactivation in cultured pea embryo axes. Effect of abscisic acid. Vitr. Cell. Dev. Biol. Anim. 1993, 29, 47–50. [Google Scholar] [CrossRef]
- Wang, H.; Qi, Q.; Schorr, P.; Cutler, A.J.; Crosby, W.L.; Fowke, L.C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both CDC2A and CYCD3, and its expression is induced by abscisic acid. Plant J. 1998, 15, 501–510. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Dhillon, S.S.; Miksche, J.P. DNA, RNA, protein and heterochromatin changes during embryo development and germination of soybean (Glycine max L.). J. Mol. Histol. 1983, 15, 21–37. [Google Scholar] [CrossRef]
- Das, S.; Pal, A. Differential DNA endoreduplication and protein profile during cotyledon ontogeny of Vigna radiata. J. Plant Biochem. Biotechnol. 2003, 12, 11–18. [Google Scholar] [CrossRef]
- Ochatt, S.; Abirached-Darmency, M. The underlying processes governing seed size plasticity: Impact of endoploidy on seed coat development and cell expansion in Medicago truncatula. In The Model Legume Medicago Truncatula; De Bruijn, F., Ed.; Wiley and Sons: Hoboken, NJ, USA, 2019; pp. 99–116. [Google Scholar]
- Ishida, T.; Adachi, S.; Yoshimura, M.; Shimizu, K.; Umeda, M.; Sugimoto, K. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 2010, 137, 63–71. [Google Scholar] [CrossRef]
- Takahashi, N.; Kajihara, T.; Okamura, C.; Kim, Y.; Katagiri, Y.; Okushima, Y.; Matsunaga, S.; Hwang, I.; Umeda, M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 2013, 23, 1812–1817. [Google Scholar] [CrossRef]
- Atif, R.M.; Boulisset, F.; Conreux, C.; Thompson, R.; Ochatt, S.J. In vitro auxin treatment promotes cell division and delays endoreduplication in developing seeds of the model legume species Medicago truncatula. Physiol. Plant 2012, 148, 549–559. [Google Scholar] [CrossRef]
- Ochatt, S.J. Agroecological impact of an in vitro biotechnology approach of embryo development and seed filling in legumes. Agron. Sustain. Dev. 2014, 35, 535–552. [Google Scholar] [CrossRef]
- Canales, C.; Bhatt, A.M.; Scott, R.; Dickinson, H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 2002, 12, 1718–1727. [Google Scholar] [CrossRef]
- Go, Y.S.; Lee, S.B.; Kim, H.J.; Kim, J.; Park, H.-Y.; Kim, J.-K.; Shibata, K.; Yokota, T.; Ohyama, K.; Muranaka, T.; et al. Identification of marneral synthase, which is critical for growth and development in Arabidopsis. Plant J. 2012, 72, 791–804. [Google Scholar] [CrossRef]
- Meinke, D.W.; Franzmann, L.H.; Nickle, T.C.; Yeung, E.C. Leafy cotyledon mutants of Arabidopsis. Plant Cell 1994, 6, 1049–1064. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Usui, H.; Hobo, T.; Takeda, S.; Hattori, T. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 2010, 51, 2031–2046. [Google Scholar] [CrossRef]
- Yamamoto, A.; Yoshii, M.; Murase, S.; Fujita, M.; Kurata, N.; Hobo, T.; Kagaya, Y.; Takeda, S.; Hattori, T. Cell-by-cell developmental transition from embryo to post-germination phase revealed by heterochronic gene expression and ER-body formation in Arabidopsis leafy cotyledon mutants. Plant Cell Physiol. 2014, 55, 2112–2125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed]
- Slater, S.M.H.; Yuan, H.Y.; Lulsdorf, M.M.; Vandenberg, A.; Zaharia, L.I.; Han, X.; Abrams, S.R. Comprehensive hormone profiling of the developing seeds of four grain legumes. Plant Cell Rep. 2013, 32, 1939–1952. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3Ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, L.; Huang, M.; He, X.; Yang, Y.; Liu, X.; Li, Y.; Hou, X. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 2018, 4, 289–298. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Nambara, E.; Naito, S.; McCourt, P. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 2004, 37, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-M.; Wheeler, S.; Xia, X.; Radchuk, R.; Weber, H.; Offler, C.E.; Patrick, J.W. Differential transcriptional networks associated with key phases of ingrowth wall construction in trans-differentiating epidermal transfer cells of Vicia faba cotyledons. BMC Plant Biol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Leviczky, T.; Molnár, E.; Papdi, C.; Őszi, E.; Horváth, G.V.; Vizler, C.; Nagy, V.; Pauk, J.; Bögre, L.; Magyar, Z. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 2019, 146, dev179333. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mohamed, D.; Dowhanik, S.; Petrella, R.; Gregis, V.; Li, J.; Wu, L.; Gazzarrini, S. Spatiotemporal restriction of FUSCA3 expression by class I BPCs promotes ovule development and coordinates embryo and endosperm growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.-A.; Floyd, S.K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 2009, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Boer, B.G.D.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Ohto, M.-A.; Fischer, R.L.; Goldberg, R.B.; Nakamura, K.; Harada, J.J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3123–3128. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, G.; Qu, L.-J.; Gu, H. Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 2008, 28, 337–346. [Google Scholar] [CrossRef]
- Du, Q.; Wang, H. Retarded embryo development 1 (RED1) regulates embryo development, seed maturation and plant growth in Arabidopsis. J. Genet. Genom. 2016, 43, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Le Gall, S.; Satour, P.; Deligny-Penninck, S.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Lalanne, D.; Pelletier, S.; Torres-Jerez, I.; Righetti, K.; Bandyopadhyay, K.; Leprince, O.; Chatelain, E.; Vu, B.L.; Gouzy, J.; et al. A Regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 2013, 163, 757–774. [Google Scholar] [CrossRef]
- Manfre, A.J.; Lanni, L.M.; Marcotte, W.R. The Arabidopsis group 1 late embryogenesis abundant protein ATEM6 is required for normal seed development. Plant Physiol. 2005, 140, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Manfre, A.J.; LaHatte, G.A.; Climer, C.R.; Marcotte, W.R. Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant ATEM6-1. Plant Cell Physiol. 2008, 50, 243–253. [Google Scholar] [CrossRef]
- Wyse, S.V.; Dickie, J.B. Predicting the global incidence of seed desiccation sensitivity. J. Ecol. 2017, 105, 1082–1093. [Google Scholar] [CrossRef]
- Dussert, S.; Serret, J.; Siqueira, A.; Morcillo, F.; Déchamp, E.; Rofidal, V.; Lashermes, P.; Etienne, H.; Joët, T. Integrative analysis of the late maturation programme and desiccation tolerance mechanisms in intermediate coffee seeds. J. Exp. Bot. 2018, 69, 1583–1597. [Google Scholar] [CrossRef]
- Chandra, J.; Keshavkant, S. Desiccation-induced ROS accumulation and lipid catabolism in recalcitrant Madhuca latifolia seeds. Physiol. Mol. Biol. Plants 2017, 24, 75–87. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Pramanik, S.K.; Bewley, J.D. The expression of dehydrin proteins in desiccation-sensitive (recalcitrant) seeds of temperate trees. Planta 1994, 193, 478–485. [Google Scholar] [CrossRef]
- Sunderlikova, V.; Salaj, J.; Kopecky, D.; Salaj, T.; Wilhem, E.; Matušíková, I. Dehydrin genes and their expression in recalcitrant oak (Quercus robur) embryos. Plant Cell Rep. 2009, 28, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Kleinwã¤Chter, M.; Radwan, A.; Hara, M.; Selmar, D. Dehydrin expression in seeds: An issue of maturation drying. Front. Plant Sci. 2014, 5, 402. [Google Scholar] [CrossRef]
- Wilhelmsson, P.K.I.; Chandler, J.O.; Fernandez-Pozo, N.; Graeber, K.; Ullrich, K.K.; Arshad, W.; Khan, S.; Hofberger, J.A.; Buchta, K.; Edger, P.P.; et al. Usability of reference-free transcriptome assemblies for detection of differential expression: A case study on Aethionema arabicum dimorphic seeds. BMC Genom. 2019, 20, 1–19. [Google Scholar] [CrossRef]
- Hong, L.; Su, W.; Zhang, Y.; Ye, C.; Shen, Y.; Li, Q.Q. Transcriptome profiling during mangrove viviparity in response to abscisic acid. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Marques, A.; Nijveen, H.; Somi, C.; Ligterink, W.; Hilhorst, H. Induction of desiccation tolerance in desiccation sensitive Citrus limon seeds. J. Integr. Plant Biol. 2019, 61, 624–638. [Google Scholar] [CrossRef]
- Mccarty, D.R.; Carson, C.B.; Stinard, P.S.; Robertson, D.S. Molecular analysis of viviparous-1: An abscisic acid-insensitive mutant of maize. Plant Cell 1989, 1, 523–532. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Yamazaki, M.; Kobayashi, M.; Hirochika, R.; Miyao, A.; Hirochika, H. Screening of the rice viviparous mutants generated by endogenous retrotransposon TOS17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 2001, 125, 1248–1257. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Xu, X.; Qu, W.; Li, J.; Xu, X.; Wang, A. Seed development and viviparous germination in one accession of a tomato rin mutant. Breed. Sci. 2016, 66, 372–380. [Google Scholar] [CrossRef]
- Delahaie, J.; Hundertmark, M.; Bove, J.; Leprince, O.; Rogniaux, H.; Buitink, J. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J. Exp. Bot. 2013, 64, 4559–4573. [Google Scholar] [CrossRef]
- Li, J.; Nie, X.; Tan, J.L.H.; Berger, F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15479–15484. [Google Scholar] [CrossRef]
- Kang, I.-H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Dhatt, B.K.; Miller, M.; Folsom, J.J.; Wang, Z.; Krassovskaya, I.; Liu, K.; Sandhu, J.; Yu, H.; Zhang, C.; et al. MADS78 and MADS79 are essential regulators of early seed development in rice. Plant Physiol. 2019, 182, 933–948. [Google Scholar] [CrossRef]
- Figueiredo, D.; Batista, R.A.; Roszak, P.J.; Hennig, L.; Köhler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 2016, 5, e20542. [Google Scholar] [CrossRef] [PubMed]
- Noguero, M.; Le Signor, C.; Vernoud, V.; Bandyopadhyay, K.; Sanchez, M.; Fu, C.; Torres-Jerez, I.; Wen, J.; Mysore, K.; Gallardo, K.; et al. DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 2015, 81, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Emery, R.N.; Ma, Q.; Atkins, C.A. The forms and sources of cytokinins in developing white lupine seeds and fruits. Plant Physiol. 2000, 123, 1593–1604. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef]
- Belmonte, M.F.; Kirkbride, R.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D.; et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444. [Google Scholar] [CrossRef]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2005, 18, 40–54. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Y.; Gao, L.; Zhao, G.; Zhou, R.; Zhang, B.; Jia, J. TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef]
- Zhao, J.; Bai, W.; Zeng, Q.; Song, S.; Zhang, M.; Li, X.; Hou, L.; Xiaoying, L.; Luo, M.; Li, D.; et al. Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2015, 67, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.-B.; Huang, H.-Y.; Hu, Y.-W.; Zhu, S.-W.; Wang, Z.-Y.; Lin, W.-H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-Y.; Nam, K.H. RAV1 negatively regulates seed development by directly repressing MINI3 and IKU2 in Arabidopsis. Mol. Cells 2018, 41, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Ingram, G.C. Family plot: The impact of the endosperm and other extra-embryonic seed tissues on angiosperm zygotic embryogenesis. F1000Research 2020, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Kondou, Y.; Nakazawa-Miklasevica, M.; Kawashima, M.; Ichikawa, T.; Yoshizumi, T.; Suzuki, K.; Ishikawa, A.; Koshi, T.; Matsui, R.; Muto, S.; et al. Retarded growth of EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 2008, 147, 1924–1935. [Google Scholar] [CrossRef]
- Yang, S.; Johnston, N.; Talideh, E.; Mitchell, S.; Jeffree, C.; Goodrich, J.; Ingram, G. The endosperm specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 2008, 135, 3501–3509. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, L.; Chen, B.; Xiong, Z.; Chen, C.; Wang, L.; Yu, J.; Lu, C.; Wei, W. Functional identification and characterization of the Brassica napus transcription factor gene BnAP2, the ortholog of Arabidopsis thaliana APETALA2. PLoS ONE 2012, 7, e33890. [Google Scholar] [CrossRef]
- Kunieda, T.; Mitsuda, N.; Ohme-Takagi, M.; Takeda, S.; Aida, M.; Tasaka, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 2008, 20, 2631–2642. [Google Scholar] [CrossRef]
- Melkus, G.; Rolletschek, H.; Radchuk, R.; Fuchs, J.; Rutten, T.; Wobus, U.; Altmann, T.; Jakob, P.; Borisjuk, L. The metabolic role of the legume endosperm: A noninvasive imaging study. Plant Physiol. 2009, 151, 1139–1154. [Google Scholar] [CrossRef]
- Kim, J.; Rudella, A.; Rodriguez, V.R.; Zybailov, B.; Olinares, P.D.B.; van Wijk, K.J. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 2009, 21, 1669–1692. [Google Scholar] [CrossRef] [PubMed]
- Myouga, F.; Motohashi, R.; Kuromori, T.; Nagata, N.; Shinozaki, K. An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J. 2006, 48, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Kovacheva, S.; Bédard, J.; Patel, R.; Dudley, P.; Twell, D.; Ríos, G.; Koncz, C.; Jarvis, P. In vivo studies on the roles of TIC110, TIC40 and HSP93 during chloroplast protein import. Plant J. 2004, 41, 412–428. [Google Scholar] [CrossRef]
- Kadirjan-Kalbach, D.K.; Yoder, D.W.; Ruckle, M.E.; Larkin, R.M.; Osteryoung, K.W. FtsHi1/ARC1is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J. 2012, 72, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Bosco, C.D.; Lezhneva, L.; Biehl, A.; Leister, D.; Strotmann, H.; Wanner, G.; Meurer, J. Inactivation of the chloroplast ATP synthase γ subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.R.; Stessman, D.J.; Spalding, M.H.; Rodermel, S.R. Alterations in photosynthesis in Arabidopsis lacking immutans, a chloroplast terminal oxidase. Photosynth. Res. 2007, 91, 11–23. [Google Scholar] [CrossRef]
- Lu, Y.; Li, C.; Wang, H.; Chen, H.; Berg, R.H.; Xia, Y. AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J. 2011, 67, 13–25. [Google Scholar] [CrossRef]
- Borisjuk, L.; Rolletschek, H.; Walenta, S.; Panitz, R.; Wobus, U.; Weber, H. Energy status and its control on embryogenesis of legumes: ATP distribution within Vicia faba embryos is developmentally regulated and correlated with photosynthetic capacity. Plant J. 2003, 36, 318–329. [Google Scholar] [CrossRef]
- Lorenz, C.; Brandt, S.; Borisjuk, L.; Rolletschek, H.; Heinzel, N.; Tohge, T.; Fernie, A.R.; Braun, H.-P.; Hildebrandt, T. The role of persulfide metabolism during Arabidopsis seed development under light and dark conditions. Front. Plant Sci. 2018, 9, 1381. [Google Scholar] [CrossRef]
- Krüßel, L.; Junemann, J.; Wirtz, M.; Birke, H.; Thornton, J.D.; Browning, L.W.; Poschet, G.; Hell, R.; Balk, J.; Braun, H.-P.; et al. The mitochondrial sulfur dioxygenase ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol. 2014, 165, 92–104. [Google Scholar] [CrossRef]
- Cordoba, J.P.; Marchetti, F.; Soto, D.; Martin, M.V.; Pagnussat, G.C.; Zabaleta, E. The CA domain of the respiratory complex I is required for normal embryogenesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 67, 1589–1603. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Brandizzi, F.; Slocombe, S.P.; El-Shami, M.; Hawes, C.; Baker, A. An Arabidopsis PEX10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003, 133, 1809–1819. [Google Scholar] [CrossRef]
- Mao, G.; Wang, R.; Guan, Y.; Liu, Y.; Zhang, S. Sulfurtransferases 1 and 2 play essential roles in embryo and seed development in Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 7548–7557. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.L.; Chen, Y.K.; Xu, X.Z.; Shen, F.; Zheng, Q.B.; Du, Z.; Wang, Y.; Wu, T.; Xu, X.F.; Han, Z.H.; et al. miR156 switches on vegetative phase change under the regulation of redox signals in apple seedlings. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zheng, Q.; Jia, X.; Chen, K.; Wang, Y.; Wu, T.; Xu, X.; Han, Z.; Zhang, Z.; Zhang, X. MdGGT1 impacts apple miR156 precursor levels via ontogenetic changes in subcellular glutathione homeostasis. Front. Plant Sci. 2019, 10, 994. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, M.; Kiegle, E.; Leo, G.; Ezquer, I. Carbohydrate reserves and seed development: An overview. Plant Reprod. 2018, 31, 263–290. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Wuillème, S.; Lemoine, R.; Kronenberger, J.; Caboche, M.; Lepiniec, L.; Rochat, C. The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. 2005, 43, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Borisjuk, L.; Walenta, S.; Weber, H.; Mueller-Klieser, W.; Wobus, U. High-resolution histographical mapping of glucose concentrations in developing cotyledons of Vicia faba in relation to mitotic activity and storage processes: Glucose as a possible developmental trigger. Plant J. 1998, 15, 583–591. [Google Scholar] [CrossRef]
- Tegeder, M.; Wang, X.-D.; Frommer, W.; Offler, C.E.; Patrick, J.W. Sucrose transport into developing seeds of Pisum sativum L. Plant J. 1999, 18, 151–161. [Google Scholar] [CrossRef]
- Wardini, T.; Wang, X.-D.; Offler, C.E.; Patrick, J.W. Induction of wall ingrowths of transfer cells occurs rapidly and depends upon gene expression in cotyledons of developing Vicia faba seeds. Protoplasma 2007, 231, 15–23. [Google Scholar] [CrossRef]
- Wardini, T.; Talbot, M.J.; Offler, C.E.; Patrick, J.W. Role of sugars in regulating transfer cell development in cotyledons of developing Vicia faba seeds. Protoplasma 2006, 230, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Borisjuk, L.; Heim, U.; Sauer, N.; Wobus, U. A role for sugar transporters during seed development: Molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell 1997, 9, 895–908. [Google Scholar] [CrossRef]
- Offler, C.E.; Liet, E.; Sutton, E.G. Transfer cell induction in cotyledons of Vicia faba L. Protoplasma 1997, 200, 51–64. [Google Scholar] [CrossRef]
- Farley, S.; Patrick, J.W.; Offler, C.E. Functional transfer cells differentiate in cultured cotyledons of Vicia faba L. seeds. Protoplasma 2000, 214, 102–117. [Google Scholar] [CrossRef]
- Tomlinson, K.L.; McHugh, S.; Labbe, H.; Grainger, J.L.; James, L.E.; Pomeroy, K.M.; Mullin, J.W.; Miller, S.S.; Dennis, D.T.; Miki, B.L.A. Evidence that the hexose-to-sucrose ratio does not control the switch to storage product accumulation in oilseeds: Analysis of tobacco seed development and effects of overexpressing apoplastic invertase. J. Exp. Bot. 2004, 55, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Morley-Smith, E.R.; Pike, M.J.; Findlay, K.; Köckenberger, W.; Hill, L.M.; Smith, A.M.; Rawsthorne, S. The transport of sugars to developing embryos is not via the bulk endosperm in oilseed rape seeds. Plant Physiol. 2008, 147, 2121–2130. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Popko, J.; Heilmann, M.; Schulmeister, S.; Dietel, K.; Schmitt, B.; Stadler, R.; Feussner, I.; Sauer, N. Sucrose transporter 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J. 2012, 73, 392–404. [Google Scholar] [CrossRef]
- Zuma, B.; Dana, M.B.; Wang, D. Prolonged expression of a putative invertase inhibitor in micropylar endosperm suppressed embryo growth in Arabidopsis. Front. Plant Sci. 2018, 9, 61. [Google Scholar] [CrossRef]
- Kaur, H.; Gupta, A.K.; Kaur, N.; Sandhu, J.S. High acid invertase activity for a prolonged period in developing seeds/podwall of wild chickpea is detrimental to seed filling. Indian J. Exp. Boil. 2012, 50, 735–743. [Google Scholar]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.S.; Pratap, A.; Gupta, S.; Sharma, K.; Tomar, R.; Singh, N.P. Physiological traits for shortening crop duration and improving productivity of greengram (Vigna radiata L. Wilczek) under high temperature. Front. Plant Sci. 2019, 10, 1508. [Google Scholar] [CrossRef] [PubMed]
- Sakr, S.; Wang, M.; Dédaldéchamp, F.; Perez-Garcia, M.-D.; Ogé, L.; Hamama, L.; Atanassova, R. The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef]
- Koornneef, M.; Léon-Kloosterziel, K.M.; Schwartz, S.H.; Zeevaart, J.A. The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol. Biochem. 1998, 36, 83–89. [Google Scholar] [CrossRef]
- Hardie, D.G.; Carling, D.; Carlson, M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998, 67, 821–855. [Google Scholar] [CrossRef]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.C.; Elias, C.; Baena-Gonzã¡lez, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Wurzinger, B.; Nukarinen, E.; Nägele, T.; Weckwerth, W.; Teige, M. The SnRK1 Kinase as central mediator of energy signaling between different organelles. Plant Physiol. 2018, 176, 1085–1094. [Google Scholar] [CrossRef]
- Radchuk, R.; Radchuk, V.; Weschke, W.; Borisjuk, L.; Weber, H. Repressing the expression of the sucrose nonfermenting-1-related protein kinase gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2005, 140, 263–278. [Google Scholar] [CrossRef]
- Radchuk, R.; Emery, R.N.; Weier, D.; Vigeolas, H.; Geigenberger, P.; Lunn, J.E.; Feil, R.; Weschke, W.; Weber, H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2009, 61, 324–338. [Google Scholar] [CrossRef]
- Chan, A.; Carianopol, C.; Tsai, A.Y.-L.; Varatharajah, K.; Chiu, R.S.; Gazzarrini, S. SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. Exp. Bot. 2017, 68, 4219–4231. [Google Scholar] [CrossRef]
- Gomez, L.D.; Gilday, A.; Feil, R.; Lunn, J.E.; Graham, I.A. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010, 64, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.; Iturriaga, G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef]
- Casey, R.; Domoney, C.; Forster, C.; Hedley, C.; Hitchin, E.; Wang, T. The effect of modifying carbohydrate metabolism on seed protein gene expression in peas. J. Plant Physiol. 1998, 152, 636–640. [Google Scholar] [CrossRef]
- Golombek, S.D.; Rolletschek, H.; Wobus, U.; Weber, H. Control of storage protein accumulation during legume seed development. J. Plant Physiol. 2001, 158, 457–464. [Google Scholar] [CrossRef]
- Vigeolas, H.; Möhlmann, T.; Martini, N.; Neuhaus, H.E.; Geigenberger, P. Embryo-specific reduction of ADP-Glc pyrophosphorylase leads to an inhibition of starch synthesis and a delay in oil accumulation in developing seeds of oilseed rape. Plant Physiol. 2004, 136, 2676–2686. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, K.; Kurt, C.; Thompson, R.; Ochatt, S. In vitro culture of immature M. truncatula grains under conditions permitting embryo development comparable to that observed in vivo. Plant Sci. 2006, 170, 1052–1058. [Google Scholar] [CrossRef]
- Rolletschek, H.; Hosein, F.; Miranda, M.; Heim, U.; Götz, K.-P.; Schlereth, A.; Borisjuk, L.; Saalbach, I.; Wobus, U.; Weber, H.; et al. Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 2005, 137, 1236–1249. [Google Scholar] [CrossRef]
- Radchuk, R.; Radchuk, V.; Götz, K.-P.; Weichert, H.; Richter, A.; Emery, R.N.; Weschke, W.; Weber, H. Ectopic expression of phosphoenolpyruvate carboxylase in Vicia narbonensis seeds: Effects of improved nutrient status on seed maturation and transcriptional regulatory networks. Plant J. 2007, 51, 819–839. [Google Scholar] [CrossRef]
- Szakonyi, D.; Byrne, M.E. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 2010, 65, 269–281. [Google Scholar] [CrossRef]
- Weijers, D.; Dijk, M.F.-V.; Vencken, R.-J.; Quint, A.; Hooykaas, P.; Offringa, R. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a ribosomal protein S5 gene. Development 2001, 128, 4289–4299. [Google Scholar] [CrossRef]
- Yan, H.; Chen, D.; Wang, Y.; Sun, Y.; Zhao, J.; Sun, M.; Peng, X. Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis. Sci. Rep. 2016, 6, 31195. [Google Scholar] [CrossRef]
- Malovichko, Y.V.; Shtark, O.Y.; Vasileva, E.N.; Nizhnikov, A.A.; Antonets, K.S. Transcriptomic insights into mechanisms of early seed maturation in the garden pea (Pisum sativum L.). Cells 2020, 9, 779. [Google Scholar] [CrossRef]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of storage proteins in plant seeds is mediated by amyloid formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef] [PubMed]
- Antonets, K.; Nizhnikov, A.A. Predicting amyloidogenic proteins in the proteomes of plants. Int. J. Mol. Sci. 2017, 18, 2155. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Nakamura, M.; Nishida, I. Defects in CTP: Phosphorylethanolamine cytidylyltransferase affect embryonic and postembryonic development in Arabidopsis. Plant Cell 2006, 18, 3370–3385. [Google Scholar] [CrossRef] [PubMed]
- Sagan, M.; Ney, B.; Duc, G. Plant symbiotic mutants as a tool to analyse nitrogen nutrition and yield relationship in field-growth peas (Pisum sativum L.). Plant Soil 1993, 153, 33–45. [Google Scholar] [CrossRef]
- New, B.; Duthion, C.; Turc, O. Phenological response of pea to water stress during reproductive development. Crop. Sci. 1994, 34, 141–146. [Google Scholar] [CrossRef]
- Munier-Jolain, N.; Larmure, A.; Salon, C. Determinism of carbon and nitrogen reserve accumulation in legume seeds. Comptes Rendus Biol. 2008, 331, 780–787. [Google Scholar] [CrossRef]
- Meckel, L.; Egli, D.B.; Phillips, R.E.; Radcliffe, D.; Leggett, J.E. Effect of moisture stress on seed growth in soybeans. Agron. J. 1984, 76, 647–650. [Google Scholar] [CrossRef]
- Egli, D.B.; Ramseur, E.L.; Zhen-Wen, Y.; Sullivan, C.H. Source-sink alterations affect the number of cells in soybean cotyledons. Crop. Sci. 1989, 29, 732–735. [Google Scholar] [CrossRef]
- Tacarindua, C.R.; Shiraiwa, T.; Homma, K.; Kumagai, E.; Sameshima, R. The response of soybean seed growth characteristics to increased temperature under near-field conditions in a temperature gradient chamber. Field Crop. Res. 2012, 131, 26–31. [Google Scholar] [CrossRef]
- Duthion, C.; Pigeaire, A. Seed lengths corresponding to the final stage in seed abortion of three grain legumes. Crop. Sci. 1991, 31, 1579–1583. [Google Scholar] [CrossRef]
- Nielsen, C.; Hall, A. Responses of cowpea (Vigna unguiculata (L.) Walp.) in the field to high night air temperature during flowering. Plant responses. Field Crop. Res. 1985, 10, 181–196. [Google Scholar] [CrossRef]
- Chakrabarti, B.; Singh, S.D.; Kumar, V.; Harit, R.C.; Misra, S. Growth and yield response of wheat and chickpea crops under high temperature. Indian J. Plant Physiol. 2013, 18, 7–14. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Bhandari, K.; Kumar, J.; Kumar, S.; Singh, S.; Siddique, K.; Nayyar, H. Impact of heat stress during seed filling on seed quality and seed yield in lentil (Lens culinaris Medikus) genotypes. J. Sci. Food Agric. 2018, 98, 5134–5141. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Sita, K.; Bhandari, K.; Kumar, S.; Kumar, J.; Prasad, P.V.; Siddique, K.; Nayyar, H. Influence of drought and heat stress, applied independently or in combination during seed development, on qualitative and quantitative aspects of seeds of lentil (Lens culinaris Medikus) genotypes, differing in drought sensitivity. Plant Cell Environ. 2018, 42, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Rice, E.K.; Martínez-Oquendo, P.; McNair, J.N. Phenology of seed maturation in babysbreath (Gypsophila paniculata) in northwest Michigan, USA, and its relation to glyphosate efficacy. Invasive Plant Sci. Manag. 2019, 12, 194–201. [Google Scholar] [CrossRef]
- Hampton, J.G.; Boelt, B.; Rolston, M.P.; Chastain, T. Effects of elevated CO2 and temperature on seed quality. J. Agric. Sci. 2012, 151, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Chalk, P.; Souza, R.; Urquiaga, S.; Alves, B.; Boddey, R. The role of arbuscular mycorrhiza in legume symbiotic performance. Soil Biol. Biochem. 2006, 38, 2944–2951. [Google Scholar] [CrossRef]
- Püschel, D.; Janoušková, M.; Voříšková, A.; Gryndlerová, H.; Vosátka, M.; Jansa, J. Arbuscular Mycorrhiza stimulates biological nitrogen fixation in two Medicago spp. through improved phosphorus acquisition. Front. Plant Sci. 2017, 8, 390. [Google Scholar] [CrossRef]
- Velázquez, E.; Carro, L.; Felix, J.D.F.; Martínez-Hidalgo, P.; Menendez, E.; Ramírez-Bahena, M.-H.; Mulas, R.; González-Andrés, F.; Martínez-Molina, E.; Peix, A. The legume nodule microbiome: A source of plant growth-promoting bacteria. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 41–70. [Google Scholar]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Doidy, J.; Zimmermann, S.D.; Wipf, D.; Courty, P.-E. Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 2016, 21, 937–950. [Google Scholar] [CrossRef]
- Ezawa, T.; Saito, K. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol. 2018, 220, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia—The roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2019, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–16. [Google Scholar] [CrossRef]
- Mamontova, T.; Afonin, A.M.; Ihling, C.; Soboleva, A.; Lukasheva, E.; Sulima, A.; Shtark, O.Y.; Akhtemova, G.A.; Povydysh, M.; Sinz, A.; et al. Profiling of seed proteome in pea (Pisum sativum L.) lines characterized with high and low responsivity to combined inoculation with nodule bacteria and arbuscular mycorrhizal fungi. Molecules 2019, 24, 1603. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Puzanskiy, R.; Avdeeva, G.S.; Yurkov, A.P.; Smolikova, G.; Yemelyanov, V.V.; Kliukova, M.S.; Shavarda, A.L.; Kirpichnikova, A.A.; Zhernakov, A.; et al. Metabolic alterations in pea leaves during arbuscular mycorrhiza development. PeerJ 2019, 7, e7495. [Google Scholar] [CrossRef]
- Kakiuchi, J.; Kamiji, Y. Relationship between phosphorus accumulation and dry matter production in soybeans. Plant Prod. Sci. 2015, 18, 344–355. [Google Scholar] [CrossRef]
- Silva, L.R.; Pereira, M.J.; Azevedo, J.; Mulas, R.; Velázquez, E.; González-Andrés, F.; Valentão, P.; Andrade, P.B. Inoculation with Bradyrhizobium japonicum enhances the organic and fatty acids content of soybean (Glycine max L. Merrill) seeds. Food Chem. 2013, 141, 3636–3648. [Google Scholar] [CrossRef]
- Sistani, N.R.; Kaul, H.-P.; Desalegn, G.; Wienkoop, S. Rhizobium impacts on seed productivity, quality, and protection of Pisum sativum upon disease stress caused by Didymella pinodes: Phenotypic, proteomic, and metabolomic traits. Front. Plant Sci. 2017, 8, 1961. [Google Scholar] [CrossRef]
- Desalegn, G.; Turetschek, R.; Wienkoop, S.; Kaul, H.-P. Didymella pinodes affects N and P uptakes and their efficiencies in a tripartite mutualism of pea. Agronomy 2019, 9, 52. [Google Scholar] [CrossRef]
- Zhukov, V.Z.; Akhtemova, G.; Zhernakov, A.; Sulima, A.; Shtark, O.; Tikhonovich, I. Evaluation of the symbiotic effectiveness of pea (Pisum sativum L.) genotypes in pot experiment. Agric. Biol. 2017, 52, 607–614. [Google Scholar] [CrossRef]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Tar’An, B.; Michaels, T.; Pauls, K. Mapping genetic factors affecting the reaction to Xanthomonas axonopodis pv. phaseoli in Phaseolus vulgaris L. under field conditions. Genome 2001, 44, 1046–1056. [Google Scholar] [CrossRef]
- Venglat, P.; Xiang, D.; Wang, E.; Datla, R. Genomics of seed development: Challenges and opportunities for genetic improvement of seed traits in crop plants. Biocatal. Agric. Biotechnol. 2014, 3, 24–30. [Google Scholar] [CrossRef]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C.; Lenhard, M. The E3 ubiquitin ligase big brother controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef]
- Adamski, N.; Anastasiou, E.; Eriksson, S.; O’Neill, C.M.; Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20115–20120. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef] [PubMed]
- Primack, R.B. Relationships among flowers, fruits, and seeds. Annu. Rev. Ecol. Syst. 1987, 18, 409–430. [Google Scholar] [CrossRef]
- Segrestin, J.; Navas, M.; Garnier, E. Reproductive phenology as a dimension of the phenotypic space in 139 plant species from the Mediterranean. New Phytol. 2019, 225, 740–753. [Google Scholar] [CrossRef]
- Dukes, J.S.; Mooney, H.A. Does global change increase the success of biological invaders? Trends Ecol. Evol. 1999, 14, 135–139. [Google Scholar] [CrossRef]
- Alpert, P.; Bone, E.; Holzapfel, C. Invasiveness, invasibility and the role of environmental stress in the spread of non-native plants. Perspect. Plant Ecol. Evol. Syst. 2000, 3, 52–66. [Google Scholar] [CrossRef]
- Lipowsky, A.; Roscher, C.; Schumacher, J.; Schmid, B. Density-independent mortality and increasing plant diversity are associated with differentiation of Taraxacum officinale into R- and K-Strategists. PLoS ONE 2012, 7, e28121. [Google Scholar] [CrossRef]
- Leishman, M.R. How well do plant traits correlate with establishment ability? Evidence from a study of 16 calcareous grassland species. New Phytol. 1999, 141, 487–496. [Google Scholar] [CrossRef]
- Kahmen, S.; Poschlod, P. Plant functional trait responses to grassland succession over 25 years. J. Veg. Sci. 2004, 15, 21–32. [Google Scholar] [CrossRef]
- Griffith, T.M.; Watson, M.A. Is evolution necessary for range expansion? Manipulating reproductive timing of a weedy annual transplanted beyond its range. Am. Nat. 2006, 167, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Sexton, P.; Boote, K.; White, J.; Peterson, C. Seed size and seed growth rate in relation to cotyledon cell volume and number in common bean. Field Crop. Res. 1997, 54, 163–172. [Google Scholar] [CrossRef]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2019, 226, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Ebisuya, M.; Briscoe, J. What does time mean in development? Development 2018, 145, dev164368. [Google Scholar] [CrossRef]
- Buendia-Monreal, M.; Gillmor, S. The times they are a-changin’: Heterochrony in plant development and evolution. Front. Plant Sci. 2018, 9, 1349. [Google Scholar] [CrossRef]
- Geuten, K.; Coenen, H. Heterochronic genes in plant evolution and development. Front. Plant Sci. 2013, 4, 381. [Google Scholar] [CrossRef]
- Li, P.; Johnston, M.O. Heterochrony in plant evolutionary studies through the twentieth century. Bot. Rev. 2000, 66, 57–88. [Google Scholar] [CrossRef]
- Takhtajan, A. Neoteny and the origin of flowering plants. In Origin and Evolution of Angiosperm; Beck, C.B., Ed.; Columbia University Press: New York, NY, USA, 1976; pp. 207–219. [Google Scholar]
- Kostyun, J.L.; Preston, J.C.; Moyle, L.C. Heterochronic developmental shifts underlie floral diversity within Jaltomata (Solanaceae). EvoDevo 2017, 8, 17. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Abe, M. Regulation of reproductive development by non-coding RNA in Arabidopsis: To flower or not to flower. J. Plant Res. 2012, 125, 693–704. [Google Scholar] [CrossRef]
- Meinke, D.W. A Homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 1992, 258, 1647–1650. [Google Scholar] [CrossRef]
- Keith, K.; Kraml, M.; Dengler, N.G.; McCourt, P. FUSCA3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 1994, 6, 589–600. [Google Scholar] [CrossRef]
- Box, M.S.; Glover, B.J. A plant developmentalist’s guide to paedomorphosis: Reintroducing a classic concept to a new generation. Trends Plant Sci. 2010, 15, 241–246. [Google Scholar] [CrossRef]
- Conway, L.J.; Poethig, R.S. Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc. Natl. Acad. Sci. USA 1997, 94, 10209–10214. [Google Scholar] [CrossRef] [PubMed]
- Gillmor, S.; Park, M.Y.; Smith, M.R.; Pepitone, R.; Kerstetter, R.A.; Poethig, R.S. The MED12–MED13 module of mediator regulates the timing of embryo patterning in Arabidopsis. Development 2010, 137, 113–122. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Wobus, U. Seed-development programs: A systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 2013, 64, 189–217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Begcy, K.; Dresselhaus, T.; Sun, M.-X. Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiol. 2016, 173, 130–142. [Google Scholar] [CrossRef]
- Chuck, G.; Candela, H.; Hake, S. Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 2009, 12, 81–86. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hakata, M. Atlas of rice grain filling-related metabolism under high temperature: Joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010, 51, 795–809. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Liu, S.-J.; Song, S.-Q.; Møller, I.M. Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Bahuguna, R.N.; Shah, D.; Pal, M.; Jagadish, S.V.K. High temperature stress during flowering and grain filling offsets beneficial impact of elevated CO2 on assimilate partitioning and sink-strength in rice. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Khakimov, B.; Rasmussen, M.A.; Kannangara, R.M.; Jespersen, B.M.; Munck, L.; Engelsen, S.B. From metabolome to phenotype: GC-MS metabolomics of developing mutant barley seeds reveals effects of growth, temperature and genotype. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Nasehzadeh, M.; Ellis, R.H. Wheat seed weight and quality differ temporally in sensitivity to warm or cool conditions during seed development and maturation. Ann. Bot. 2017, 120, 479–493. [Google Scholar] [CrossRef]
- Begcy, K.; Sandhu, J.; Walia, H. Transient heat stress during early seed development primes germination and seedling establishment in rice. Front. Plant Sci. 2018, 9, 1768. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, L.; Lu, Y.; Wu, B.; Gong, X.; Liu, M.; Wang, J.; Sun, Q.; Vierling, E.; Xu, S. Metabolic adaptation of wheat grains contributes to a stable filling rate under heat stress. J. Exp. Bot. 2018, 69, 5531–5545. [Google Scholar] [CrossRef]
- Boehlein, S.K.; Liu, P.; Webster, A.; Ribeiro, C.; Suzuki, M.; Wu, S.; Guan, J.; Stewart, J.D.; Tracy, W.F.; Settles, A.M.; et al. Effects of long-term exposure to elevated temperature on Zea mays endosperm development during grain fill. Plant J. 2019, 99, 23–40. [Google Scholar] [CrossRef]
- Li, M.; Kennedy, A.; Huybrechts, M.; Dochy, N.; Geuten, K. The effect of ambient temperature on Brachypodium distachyon development. Front. Plant Sci. 2019, 10, 1011. [Google Scholar] [CrossRef]
- Begcy, K.; Walia, H. Drought stress delays endosperm development and misregulates genes associated with cytoskeleton organization and grain quality proteins in developing wheat seeds. Plant Sci. 2015, 240, 109–119. [Google Scholar] [CrossRef]
- Bona, E.; Scarafoni, A.; Marsano, F.; Boatti, L.; Copetta, A.; Massa, N.; Gamalero, E.; D’Agostino, G.; Cesaro, P.; Cavaletto, M.; et al. Arbuscular mycorrhizal symbiosis affects the grain proteome of Zea mays: A field study. Sci. Rep. 2016, 6, 26439. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malovichko, Y.V.; Shikov, A.E.; Nizhnikov, A.A.; Antonets, K.S. Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications. Int. J. Mol. Sci. 2021, 22, 9252. https://doi.org/10.3390/ijms22179252
Malovichko YV, Shikov AE, Nizhnikov AA, Antonets KS. Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications. International Journal of Molecular Sciences. 2021; 22(17):9252. https://doi.org/10.3390/ijms22179252
Chicago/Turabian StyleMalovichko, Yury V., Anton E. Shikov, Anton A. Nizhnikov, and Kirill S. Antonets. 2021. "Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications" International Journal of Molecular Sciences 22, no. 17: 9252. https://doi.org/10.3390/ijms22179252
APA StyleMalovichko, Y. V., Shikov, A. E., Nizhnikov, A. A., & Antonets, K. S. (2021). Temporal Control of Seed Development in Dicots: Molecular Bases, Ecological Impact and Possible Evolutionary Ramifications. International Journal of Molecular Sciences, 22(17), 9252. https://doi.org/10.3390/ijms22179252






