High Resolution Analysis of Proteome Dynamics during Bacillus subtilis Sporulation
Abstract
:1. Introduction
2. Results
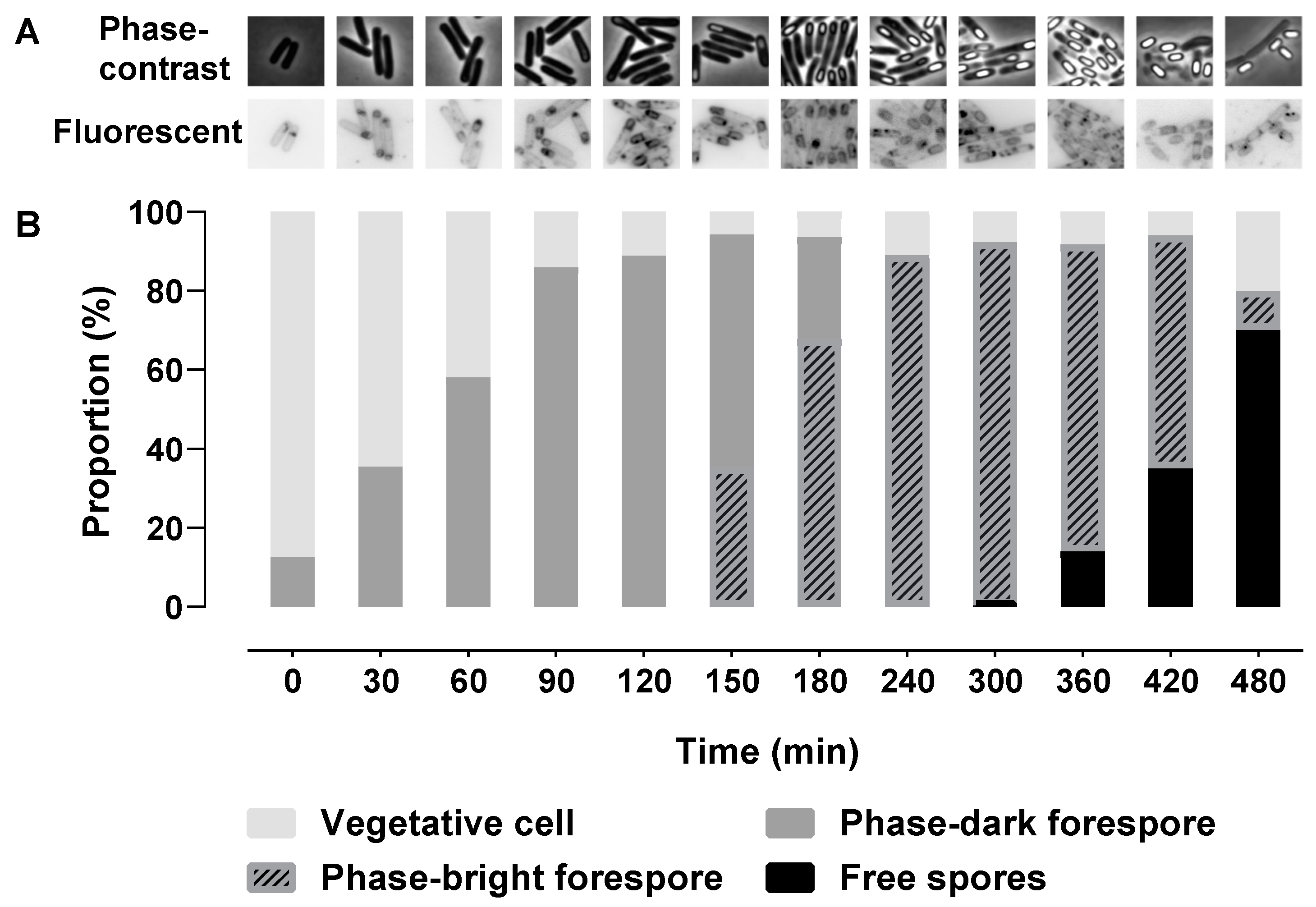
2.1. Morphological Stages during Sporulation
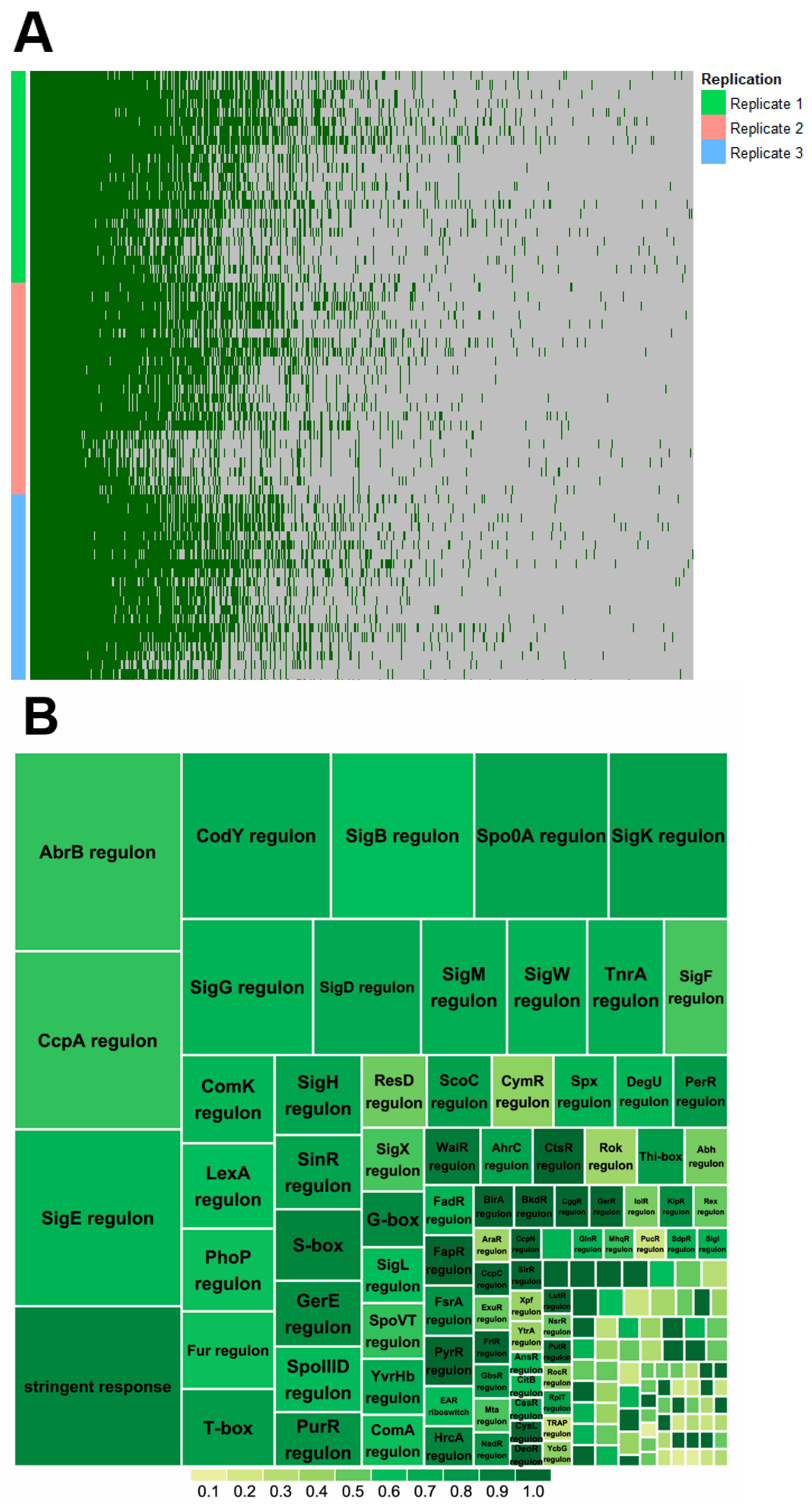
2.2. Proteome Coverage of Sporulation
2.3. Sporulation Regulatory Proteins
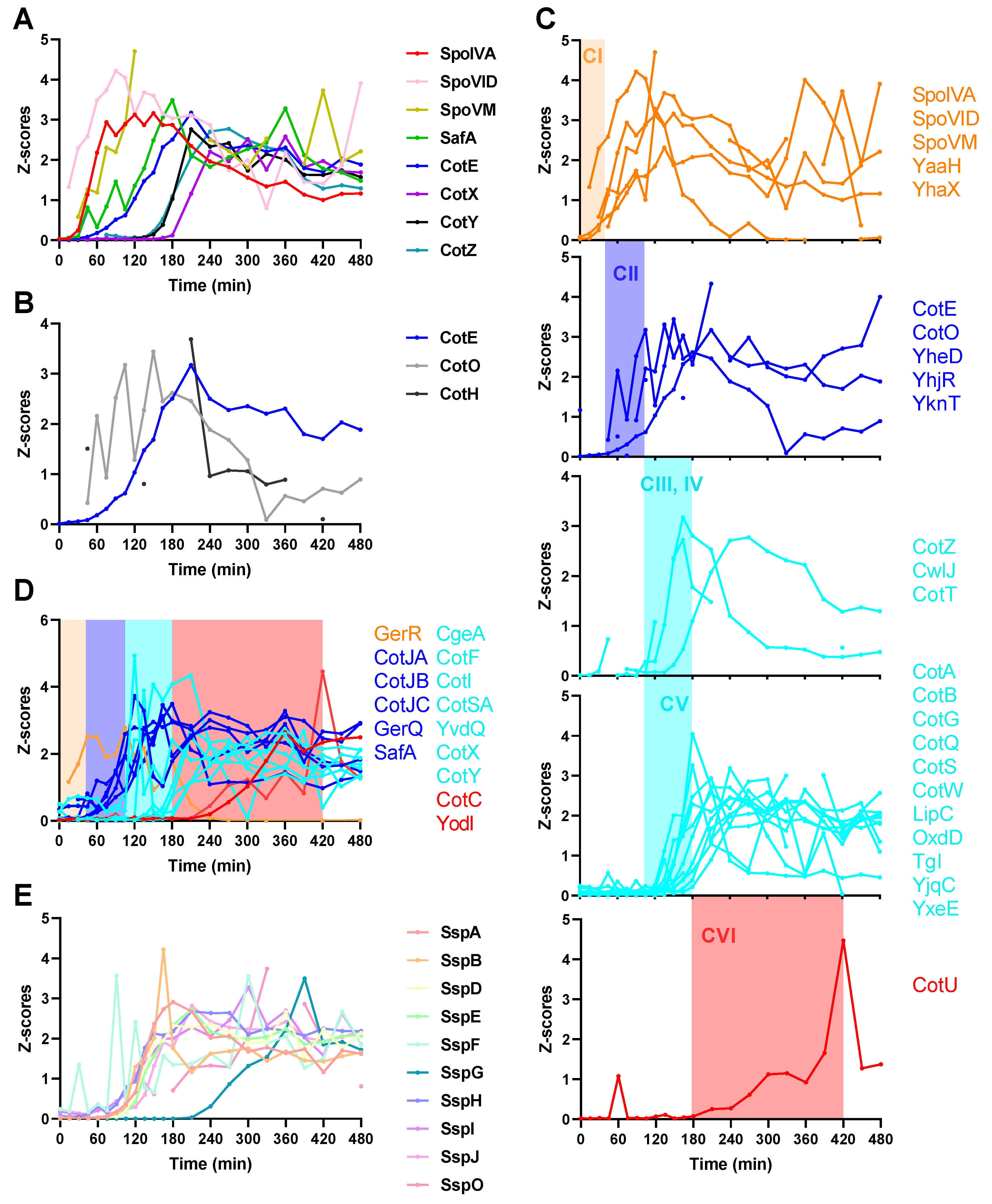
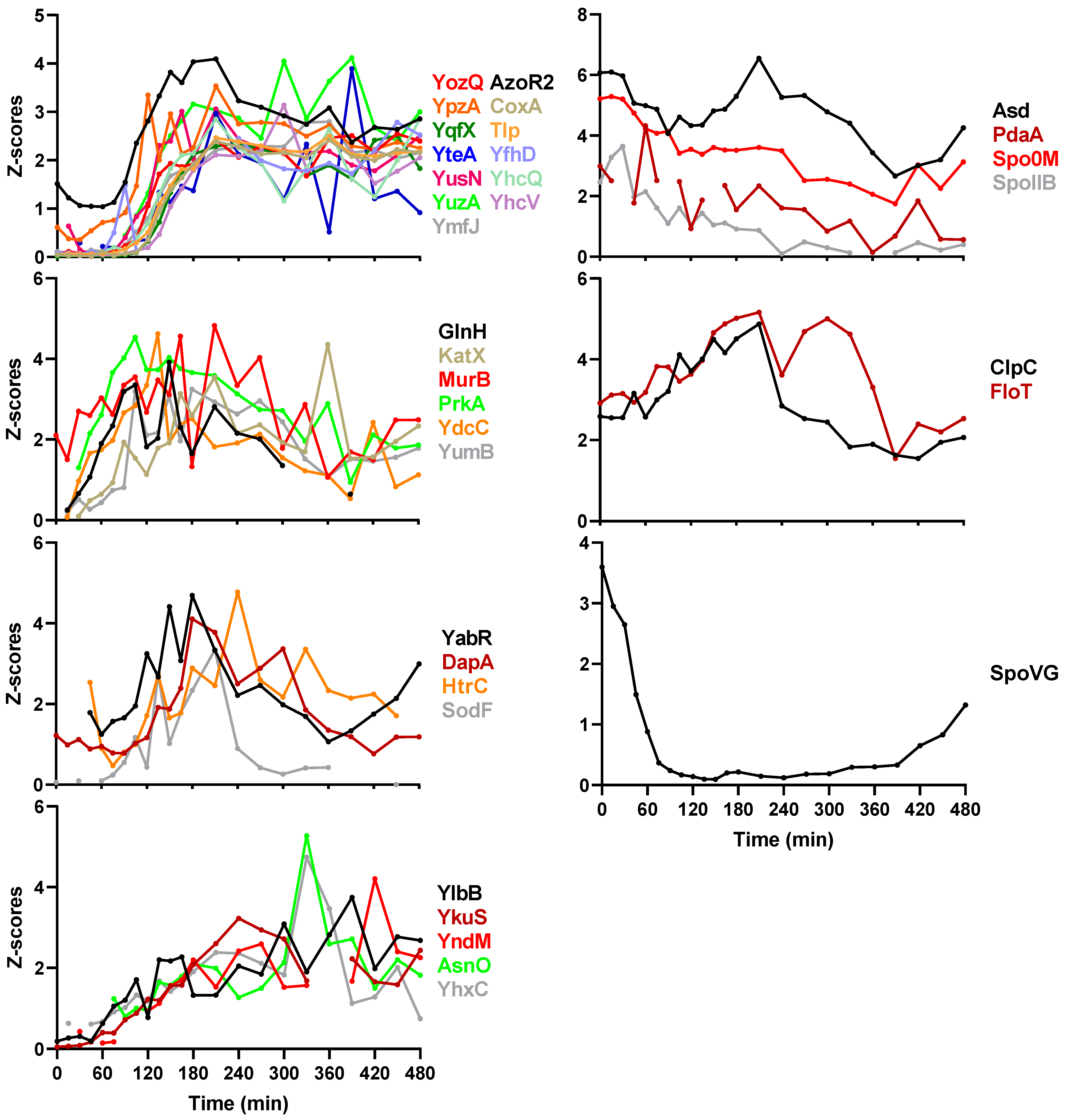
2.4. Spore Coat Proteins, Small Acid-Soluble Spore Proteins (SASPs), and Other Sporulation Proteins
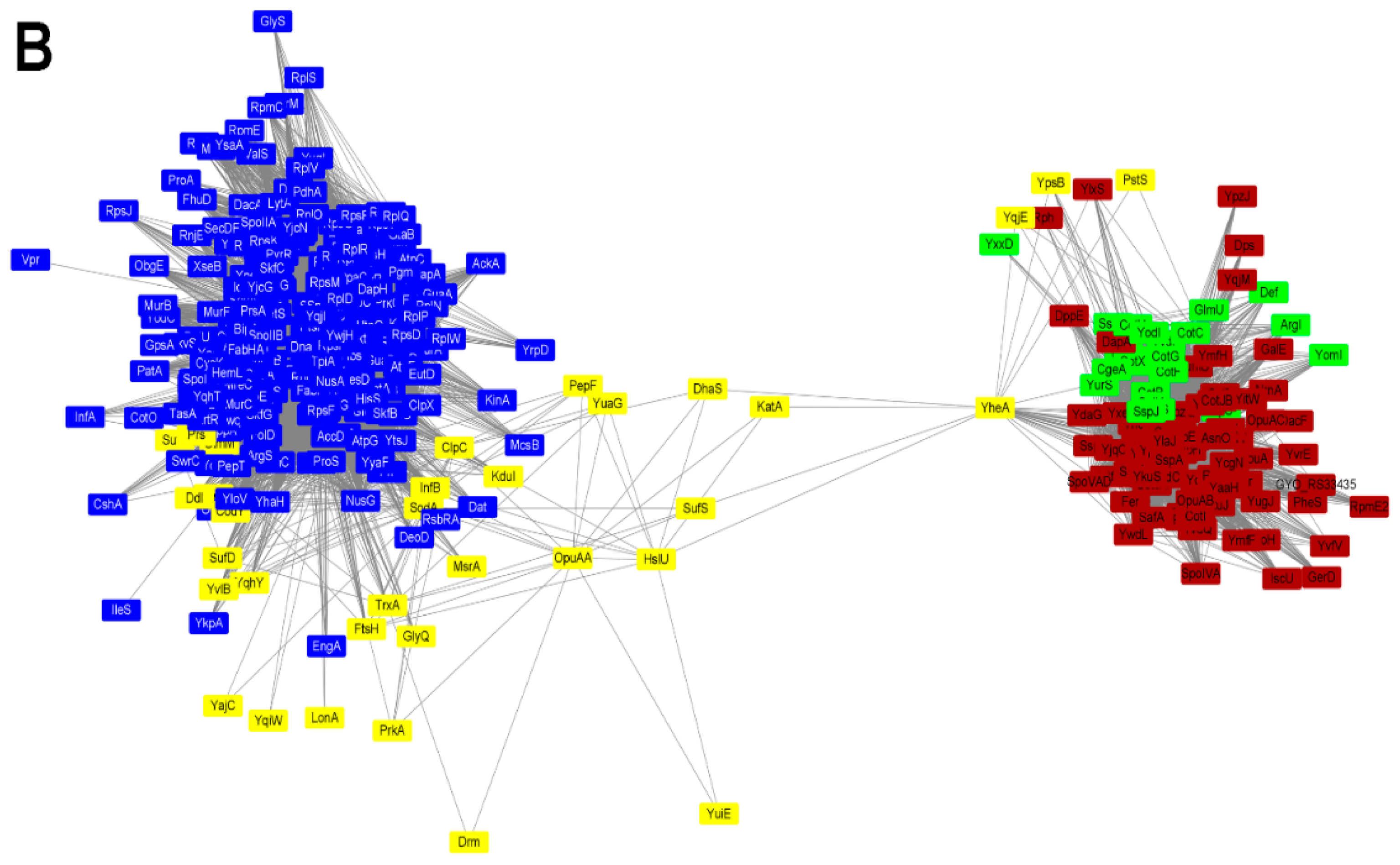
2.5. Protein Co-Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Sporulation and Sampling
4.2. Microscopy
4.3. LC-MS/MS
4.4. Data Processing and Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, I.S.; Ramamurthi, K.S. Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 2014, 6, 212–225. [Google Scholar] [CrossRef] [Green Version]
- McKenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013, 11, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Errington, J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003, 1, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.; Dworkin, J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012, 36, 131–148. [Google Scholar] [CrossRef] [Green Version]
- Henriques, A.O.; Moran, C.P. Structure, sssembly, and function of the spore surface layers. Annu. Rev. Microbiol. 2007, 61, 555–588. [Google Scholar] [CrossRef]
- De Hoon, M.J.; Eichenberger, P.; Vitkup, D. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 2010, 20, R735–R745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camp, A.H.; Losick, R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009, 23, 1014–1024. [Google Scholar] [CrossRef] [Green Version]
- Doan, T.; Morlot, C.; Meisner, J.; Serrano, M.; Henriques, A.O.; Moran, C.P., Jr.; Rudner, D.Z. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009, 5, e1000566. [Google Scholar] [CrossRef] [Green Version]
- Tu, Z.; Abhyankar, W.; Swarge, B.; van der Wel, N.; Kramer, G.; Brul, S.; de Koning, L. Artificial sporulation induction (ASI) by kinA overexpression affects the proteomes and properties of Bacillus subtilis spores. Int. J. Mol. Sci. 2020, 21, 4315. [Google Scholar] [CrossRef]
- Swarge, B.; Abhyankar, W.; Jonker, M.; Hoefsloot, H.; Kramer, G.; Setlow, P.; Brul, S.; de Koning, L.J. Integrative analysis of proteome and transcriptome dynamics during Bacillus subtilis spore revival. mSphere 2020, 5, e00463-20. [Google Scholar] [CrossRef]
- Omony, J.; de Jong, A.; Krawczyk, A.O.; Eijlander, R.T.; Kuipers, O.P. Dynamic sporulation gene co-expression networks for Bacillus subtilis 168 and the food-borne isolate Bacillus amyloliquefaciens: A transcriptomic model. Microb. Genom. 2018, 4, e000157. [Google Scholar] [CrossRef] [Green Version]
- Arrieta-Ortiz, M.L.; Hafemeister, C.; Bate, A.R.; Chu, T.; Greenfield, A.; Shuster, B.; Barry, S.N.; Gallitto, M.; Liu, B.; Kacmarczyk, T. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol. Syst. Biol. 2015, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Eswaramoorthy, P.; Duan, D.; Dinh, J.; Dravis, A.; Devi, S.N.; Fujita, M. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J. Bacteriol. 2010, 192, 3870–3882. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Stülke, J. SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 2018, 46, D743–D748. [Google Scholar] [CrossRef] [PubMed]
- Meisner, J.; Wang, X.; Serrano, M.; Henriques, A.O.; Moran, C.P. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. USA 2008, 105, 15100–15105. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Hahn, M.; Grabowski, P.; McPherson, D.C.; Otte, M.M.; Wang, R.; Ferguson, C.C.; Eichenberger, P.; Driks, A. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 2006, 59, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Driks, A.; Roels, S.; Beall, B.; Moran, C.P.; Losick, R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994, 8, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramamurthi, K.S.; Clapham, K.R.; Losick, R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 2006, 62, 1547–1557. [Google Scholar] [CrossRef]
- Levin, P.A.; Fan, N.; Ricca, E.; Driks, A.; Losick, R.; Cutting, S. An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 1993, 9, 761–771. [Google Scholar] [CrossRef]
- Roels, S.; Driks, A.; Losick, R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 1992, 174, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Takamatsu, H.; Kodama, T.; Nakayama, T.; Watabe, K. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 1999, 181, 4986–4994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.B.; Donovan, W.P.; Fitz-James, P.C.; Losick, R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988, 2, 1047–1054. [Google Scholar] [CrossRef] [Green Version]
- McKenney, P.T.; Driks, A.; Eskandarian, H.A.; Grabowski, P.; Guberman, J.; Wang, K.H.; Gitai, Z.; Eichenberger, P. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr. Biol. 2010, 20, 934–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, D.; Kuwana, R.; Takamatsu, H.; Watabe, K. Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J. Bacteriol. 2011, 193, 4075–4080. [Google Scholar] [CrossRef] [Green Version]
- McPherson, D.C.; Kim, H.; Hahn, M.; Wang, R.; Grabowski, P.; Eichenberger, P.; Driks, A. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 2005, 187, 8278–8290. [Google Scholar] [CrossRef] [Green Version]
- Naclerio, G.; Baccigalupi, L.; Zilhao, R.; De Felice, M.; Ricca, E. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 1996, 178, 4375–4380. [Google Scholar] [CrossRef] [Green Version]
- McKenney, P.T.; Eichenberger, P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol. Microbiol. 2012, 83, 245–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Melly, E.; Genest, P.C.; Gilmore, M.E.; Little, S.; Popham, D.L.; Driks, A.; Setlow, P. Analysis of the properties of spores of Bacillus subtilis prepared at different temperatures. J. Appl. Microbiol. 2002, 92, 1105–1115. [Google Scholar] [CrossRef]
- Isticato, R.; Lanzilli, M.; Petrillo, C.; Donadio, G.; Baccigalupi, L.; Ricca, E. Bacillus subtilis builds structurally and functionally different spores in response to the temperature of growth. Environ. Microbiol. 2020, 22, 170–182. [Google Scholar] [CrossRef]
- Abhyankar, W.R.; Kamphorst, K.; Swarge, B.N.; van Veen, H.; van der Wel, N.N.; Brul, S.; de Koster, C.G.; de Koning, L.J. The influence of sporulation conditions on the spore coat protein composition of Bacillus subtilis spores. Front. Microbiol. 2016, 7, 1636. [Google Scholar] [CrossRef] [Green Version]
- Bergman, N.H.; Anderson, E.C.; Swenson, E.E.; Niemeyer, M.M.; Miyoshi, A.D.; Hanna, P.C. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J. Bacteriol. 2006, 188, 6092–6100. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Ter Beek, A.; Vischer, N.O.; Smelt, J.P.; Brul, S.; Manders, E.M. Live cell imaging of germination and outgrowth of individual Bacillus subtilis spores; the effect of heat stress quantitatively analyzed with SporeTracker. PLoS ONE 2013, 8, e58972. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Abhyankar, W.; Stelder, S.; de Koning, L.; de Koster, C.; Brul, S. ‘Omics’ for microbial food stability: Proteomics for the development of predictive models for bacterial spore stress survival and outgrowth. Int. J. Food Microbiol. 2017, 240, 11–18. [Google Scholar] [CrossRef]
- Ghosh, S.; Korza, G.; Maciejewski, M.; Setlow, P. Analysis of metabolism in dormant spores of Bacillus species by 31P nuclear magnetic resonance analysis of low-molecular-weight compounds. J. Bacteriol. 2015, 197, 992–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarge, B.N.; Roseboom, W.; Zheng, L.; Abhyankar, W.R.; Brul, S.; de Koster, C.G.; de Koning, L.J. “One-pot” sample processing method for proteome-wide analysis of microbial cells and spores. Proteom. Clin. Appl. 2018, 12, 1700169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scopes, R.K. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 1974, 59, 277–282. [Google Scholar] [CrossRef]
- Honaker, J.; King, G.; Blackwell, M. Amelia II: A program for missing data. J. Stat. Soft. 2011, 45, 1–47. [Google Scholar] [CrossRef]


| Module | UniProt ID | Protein | Function | Product |
|---|---|---|---|---|
| Brown | P50727 | Fer | Electron transfer | Ferredoxin |
| Brown | O31796 | Hfq | Unknown | RNA chaperone |
| Brown | O07609 | YhfK | Unknown | Unknown |
| Brown | P45872 | PrfA | Translation | Peptide chain release factor 1 |
| Brown | P94521 | YsdC | Unknown | Unknown |
| Brown | P54550 | YqjM | Reduction of double bonds of nonsaturated aldehydes | NADPH-dependent flavin oxidoreductase |
| Brown | O34389 | MalS | Malate utilization | Malate dehydrogenase (decarboxylating) |
| Brown | P28619 | Rph | 3–5 exoribonuclease | RNase PH (EC 2.7.7.56) |
| Brown | O34503 | YtzD | Unknown | Unknown |
| Brown | P94425 | YcnE | Unknown | Unknown |
| Brown | O31509 | YeeI | Unknown | Unknown |
| Brown | C0H447 | YpzJ | Unknown | Unknown |
| Brown | O34600 | NrnA | Degradation of RNA oligonucleotides | Oligoribonuclease (nanoRNase), 3,5-bisphosphate nucleotidase |
| Brown | P46343 | PhoH | Unknown | Unknown |
| Brown | P70949 | YitW | Assembly of iron-sulfur clusters | Iron-sulfur cluster assembly factor |
| Green | P49778 | Efp | Translation | Elongation factor P |
| Green | Q45495 | Def | Utilization of S-methyl-cysteine | N-formylcysteine deformylase |
| Green | P54457 | YqeL | Translation | Ribosomal silencing factor |
| Green | P40737 | YxxD | Inhibition of the cytotoxic activity of YxiD | Antitoxin |
| Green | O31976 | YomI | Cell wall turnover | Cell wall hydrolase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, Z.; Dekker, H.L.; Roseboom, W.; Swarge, B.N.; Setlow, P.; Brul, S.; Kramer, G. High Resolution Analysis of Proteome Dynamics during Bacillus subtilis Sporulation. Int. J. Mol. Sci. 2021, 22, 9345. https://doi.org/10.3390/ijms22179345
Tu Z, Dekker HL, Roseboom W, Swarge BN, Setlow P, Brul S, Kramer G. High Resolution Analysis of Proteome Dynamics during Bacillus subtilis Sporulation. International Journal of Molecular Sciences. 2021; 22(17):9345. https://doi.org/10.3390/ijms22179345
Chicago/Turabian StyleTu, Zhiwei, Henk L. Dekker, Winfried Roseboom, Bhagyashree N. Swarge, Peter Setlow, Stanley Brul, and Gertjan Kramer. 2021. "High Resolution Analysis of Proteome Dynamics during Bacillus subtilis Sporulation" International Journal of Molecular Sciences 22, no. 17: 9345. https://doi.org/10.3390/ijms22179345
APA StyleTu, Z., Dekker, H. L., Roseboom, W., Swarge, B. N., Setlow, P., Brul, S., & Kramer, G. (2021). High Resolution Analysis of Proteome Dynamics during Bacillus subtilis Sporulation. International Journal of Molecular Sciences, 22(17), 9345. https://doi.org/10.3390/ijms22179345








