Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis
Abstract
1. Introduction
2. Results
2.1. Identification and Annotation of MADS-Box Genes in A. amurensis
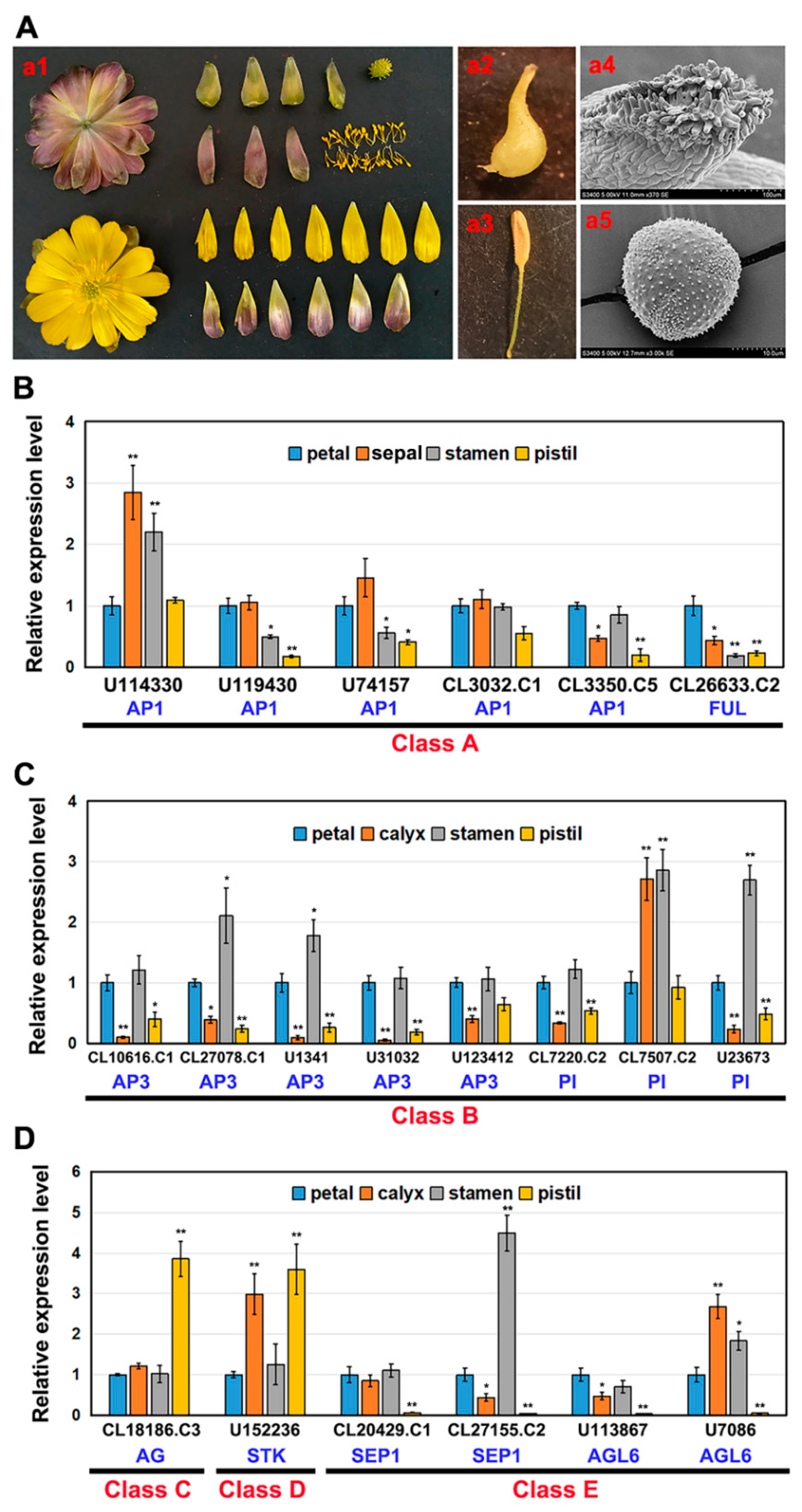
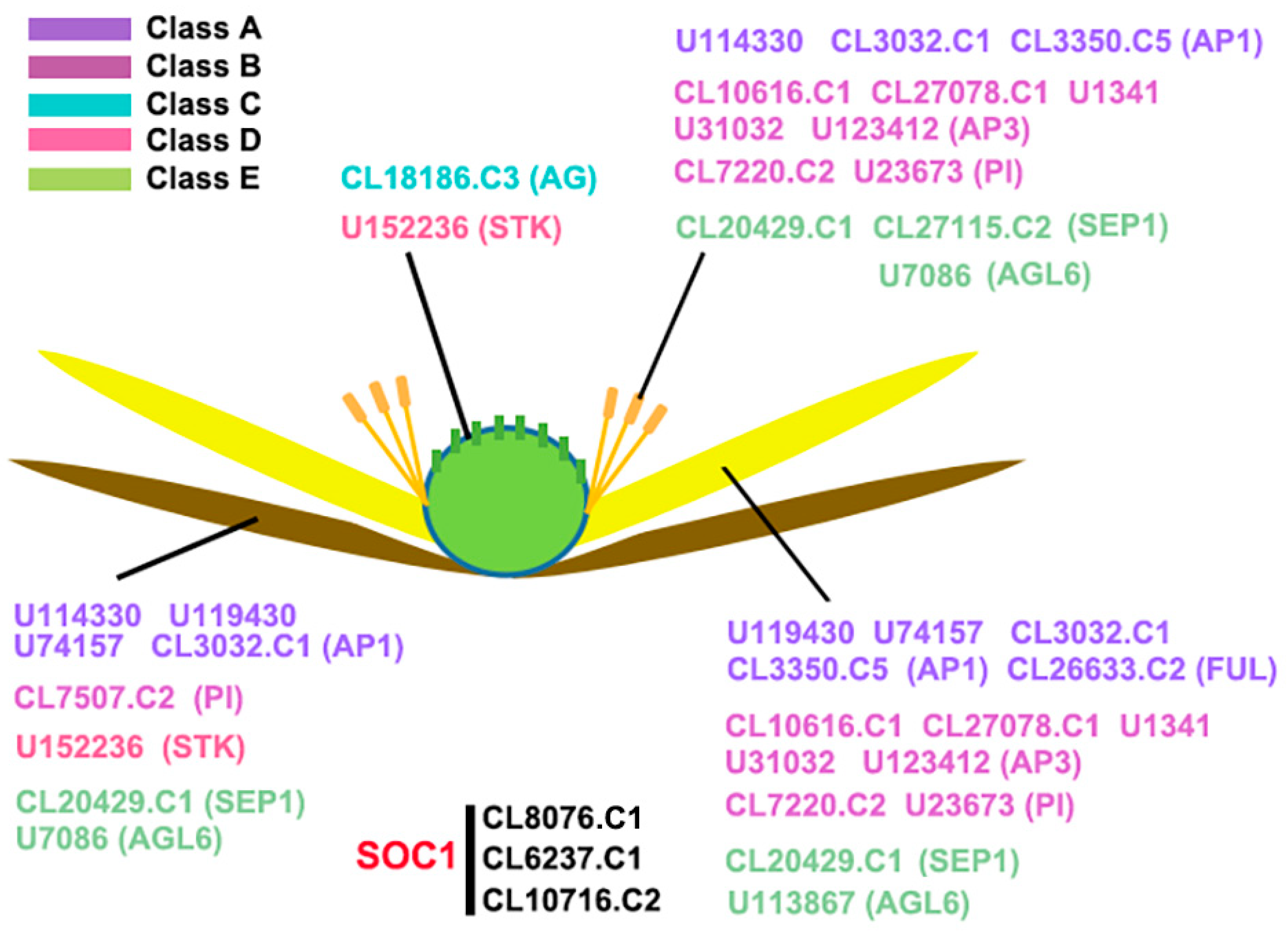
2.2. Expression of AaMADS Genes in the Flowers of A. amurensis
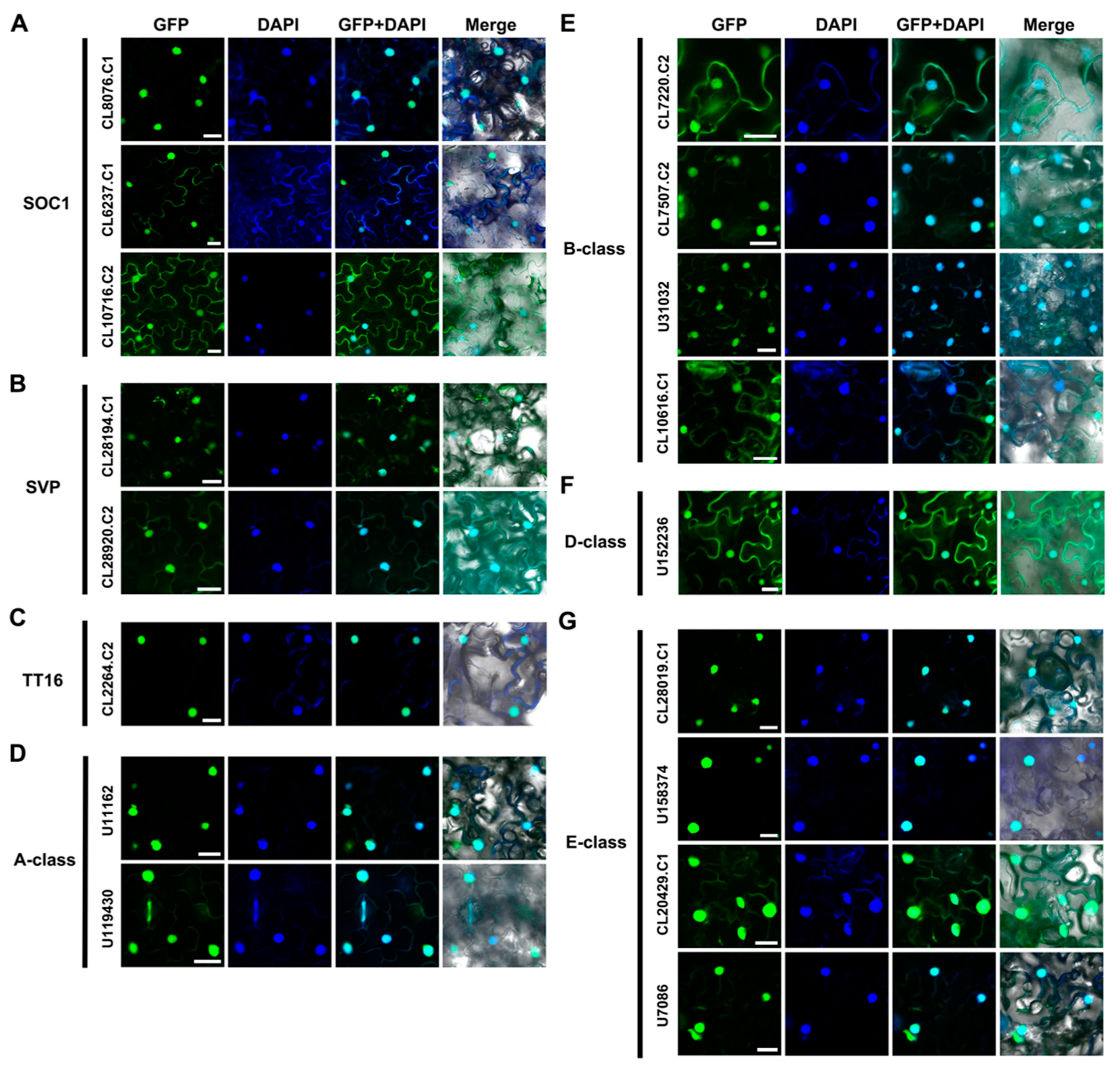
2.3. Subcellular Localization of AaMADS Proteins
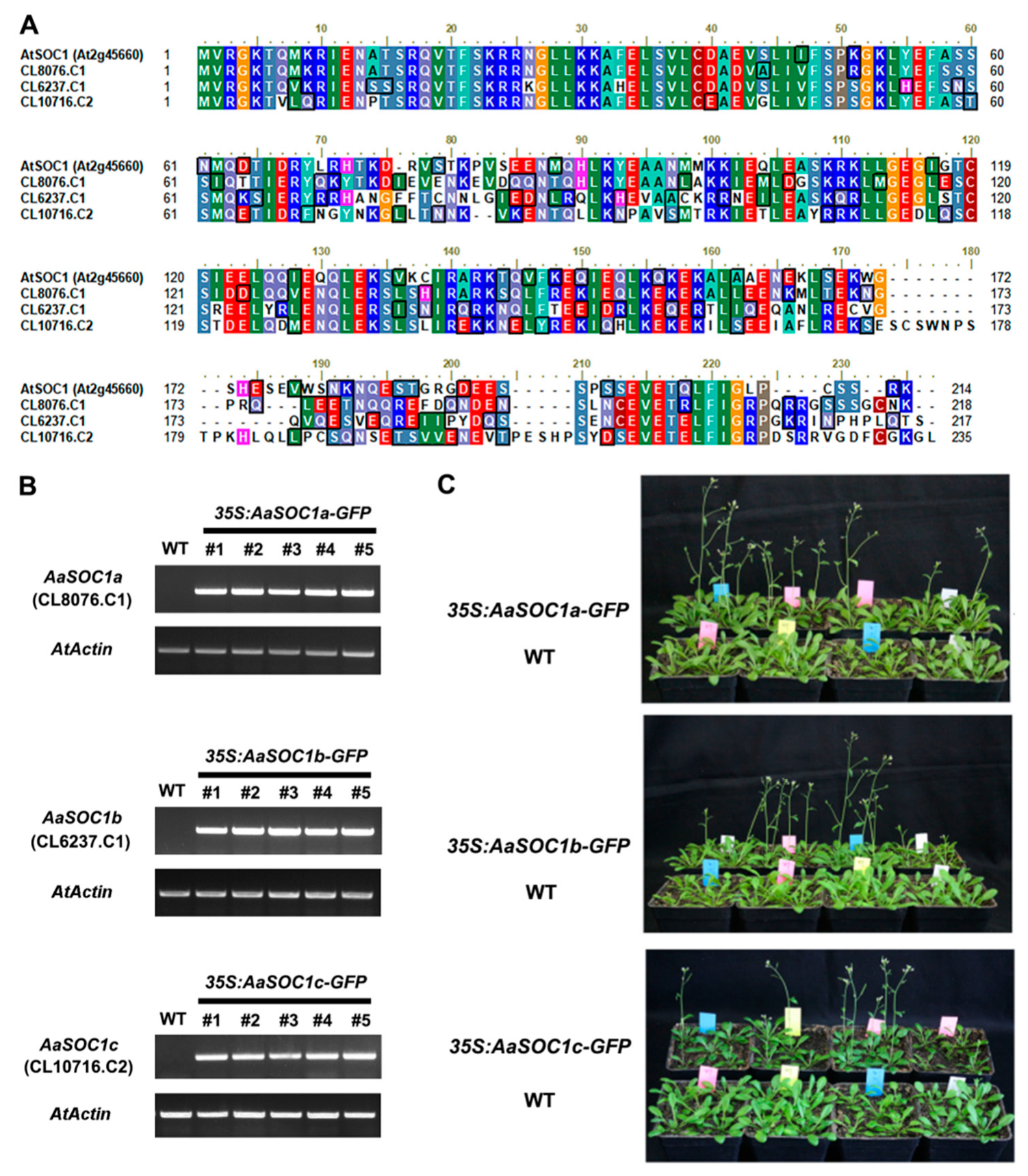
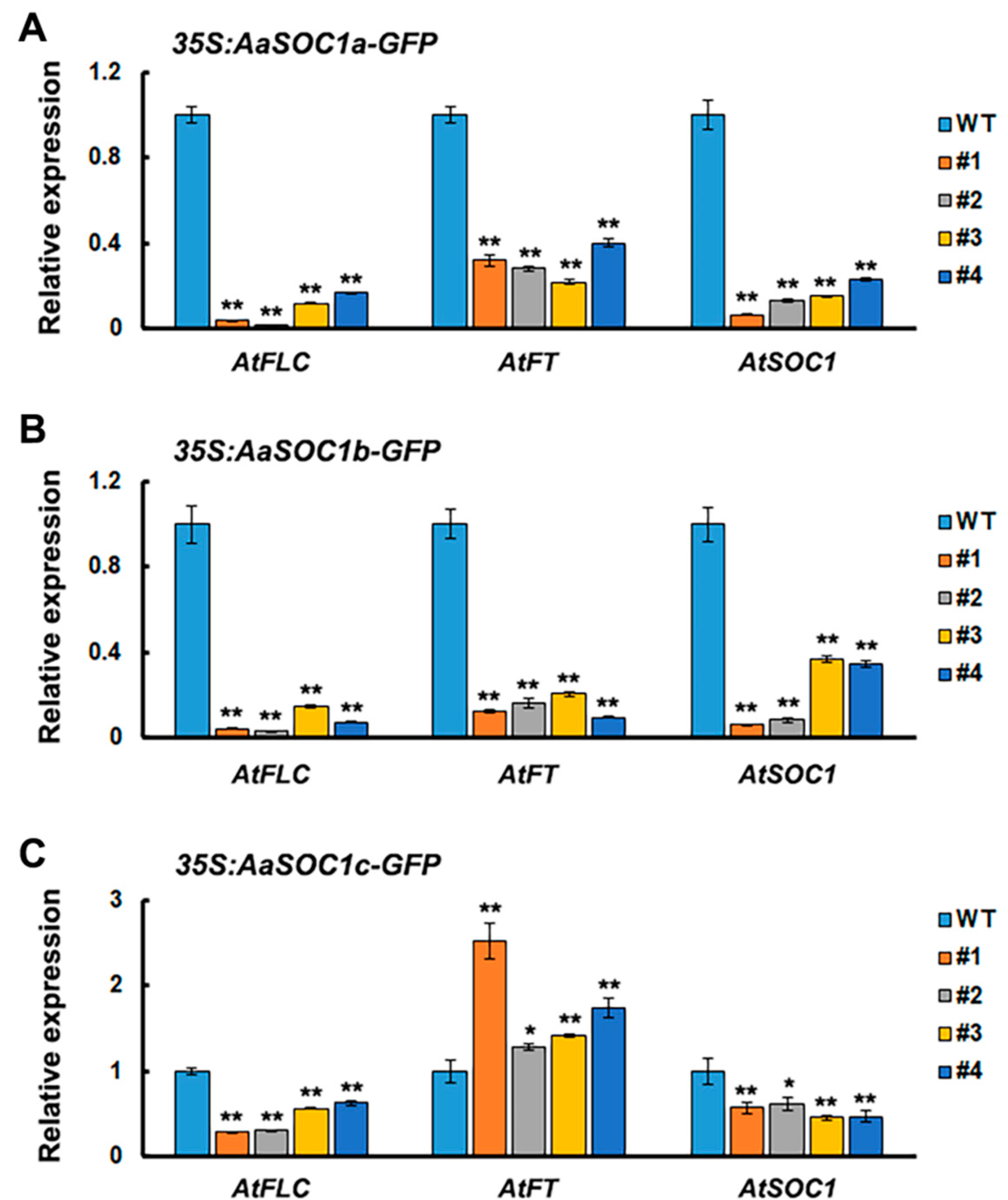
2.4. Characterization of Three SOC1-Type AaMADS Genes
3. Discussion
4. Materials and Methods
4.1. Identification of MADS-Box Genes in A. amurensis
4.2. Conserved Motifs and Phylogenetic Analysis of MADS-Box Proteins
4.3. Expression-Pattern Clustering of AaMADS Genes
4.4. Quantitative Real-Time PCR (qPCR) Analysis
4.5. Gene Cloning, Vector Construction, and Plant Transformation
4.6. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Messenguy, F.; Dubois, E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 2003, 316, 1–21. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Jack, T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.H.; Ma, H.; Nei, M.S. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef]
- Hartmann, U.; Hohmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.Y.; Chen, W.J.; Peng, X.J.; Cheng, X.A.; Zhu, S.W.; Cheng, B.J. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tissue Organ Cult. 2011, 105, 159–173. [Google Scholar] [CrossRef]
- Shu, Y.J.; Yu, D.S.; Wang, D.; Guo, D.L.; Guo, C.H. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.S.; Chen, Y.L.; Liang, D.Y.; Sun, X.C.; Karanja, B.K.; Luo, X.B.; Liu, L.W. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, D.; Lin, X.; Ding, M.; Tong, Z. Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol. 2018, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, J.; Bao, X.; Wu, Q.; Yang, T.; Li, H.; Wang, W.; Zhang, Y.; Bai, N.; Guan, Y.; et al. Genome-wide analysis of Jatropha curcas MADS-box gene family and functional characterization of the JcMADS40 gene in transgenic rice. BMC Genom. 2020, 21, 325. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.Y.; Lin, L.F.; Lin, M.D.; Hsieh, S.C.; Li, A.Y.S.; Tsay, Y.S.; Chou, M.L. Overexpression of Lilium formosanum MADS-box (LFMADS) causing floral defects while promoting flowering in Arabidopsis thaliana, whereas only affecting floral transition time in Nicotiana tabacum. Int. J. Mol. Sci. 2018, 19, 2217. [Google Scholar] [CrossRef]
- Liu, J.; Fu, X.; Dong, Y.; Lu, J.; Ren, M.; Zhou, N.; Wang, C. MIKC(C)-type MADS-box genes in Rosa chinensis: The remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic. Res. 2018, 5, 25. [Google Scholar] [CrossRef]
- Zhou, A.; Sun, H.; Dai, S.; Feng, S.; Zhang, J.; Gong, S.; Wang, J. Identification of transcription factors involved in the regulation of flowering in Adonis amurensis through combined RNA-seq transcriptomics and iTRAQ proteomics. Genes 2019, 10, 305. [Google Scholar] [CrossRef]
- Zhang, X.N.; Wang, Q.J.; Yang, S.Z.; Lin, S.N.; Bao, M.Z.; Bendahmane, M.; Wu, Q.S.; Wang, C.Y.; Fu, X.P. Identification and characterization of the MADS-box genes and their contribution to flower organ in carnation (Dianthus caryophyllus L.). Genes 2018, 9, 193. [Google Scholar] [CrossRef]
- Sharma, B.; Kramer, E.M. The mads-box gene family of the basal eudicot and hybrid Aquilegia coerulea ‘Origami’ (Ranunculaceae). Ann. Mo. Bot. Gard. 2014, 99, 313–322. [Google Scholar] [CrossRef][Green Version]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550–550.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.Z.; Luo, Y.Z.; Zhang, Z.L.; Xu, M.Y.; Wang, W.B.; Zhao, Y.M.; Zhang, L.; Fan, Y.L.; Wang, L. ZmSOC1, an MADS-box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int. J. Mol. Sci. 2014, 15, 19987–20003. [Google Scholar] [CrossRef]
- Liu, S.N.; Ma, T.F.; Ma, L.Y.; Lin, X.C. Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis and rice. Acta Physiol. Plant 2016, 38, 166. [Google Scholar] [CrossRef]
- Liu, X.R.; Pan, T.; Liang, W.Q.; Gao, L.; Wang, X.J.; Li, H.Q.; Liang, S. Overexpression of an orchid (Dendrobium nobile) SOC1/TM3-like ortholog, DnAGL19, in Arabidopsis regulates HOS1-FT expression. Front. Plant Sci. 2016, 7, 99. [Google Scholar] [CrossRef]
- Gui, J.; Zheng, S.; Shen, J.; Li, L. Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 2015, 6, 804. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Sun, H.; Dai, S.; Feng, S.; Qiao, K.; Wang, J.; Gong, S.; Zhou, A. Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis. Int. J. Mol. Sci. 2021, 22, 9362. https://doi.org/10.3390/ijms22179362
Ren L, Sun H, Dai S, Feng S, Qiao K, Wang J, Gong S, Zhou A. Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis. International Journal of Molecular Sciences. 2021; 22(17):9362. https://doi.org/10.3390/ijms22179362
Chicago/Turabian StyleRen, Lulu, Hongwei Sun, Shengyue Dai, Shuang Feng, Kun Qiao, Jingang Wang, Shufang Gong, and Aimin Zhou. 2021. "Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis" International Journal of Molecular Sciences 22, no. 17: 9362. https://doi.org/10.3390/ijms22179362
APA StyleRen, L., Sun, H., Dai, S., Feng, S., Qiao, K., Wang, J., Gong, S., & Zhou, A. (2021). Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis. International Journal of Molecular Sciences, 22(17), 9362. https://doi.org/10.3390/ijms22179362






