Charge-Transfer Interactions Stabilize G-Quadruplex-Forming Thrombin Binding Aptamers and Can Improve Their Anticoagulant Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Solid Supported Synthesis of NDI and DAN TBA Analogues and Their Characterization
2.2. NDI and DAN Ends Confer Higher Thermal Stability to TBA
2.2.1. Conformational and Spectroscopic Properties of the Modified TBA Analogues
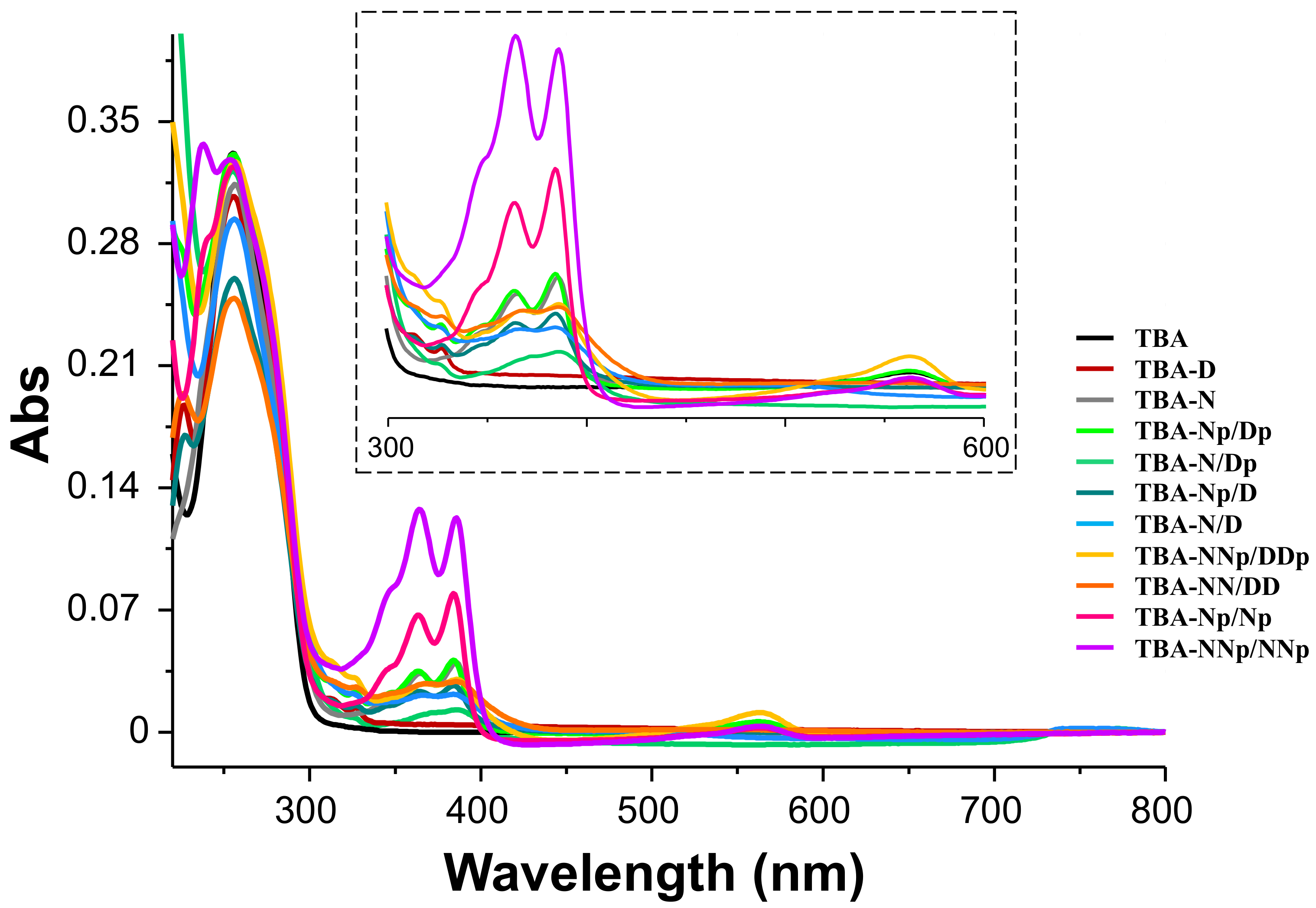
2.2.1.1. UV Spectroscopy Analysis
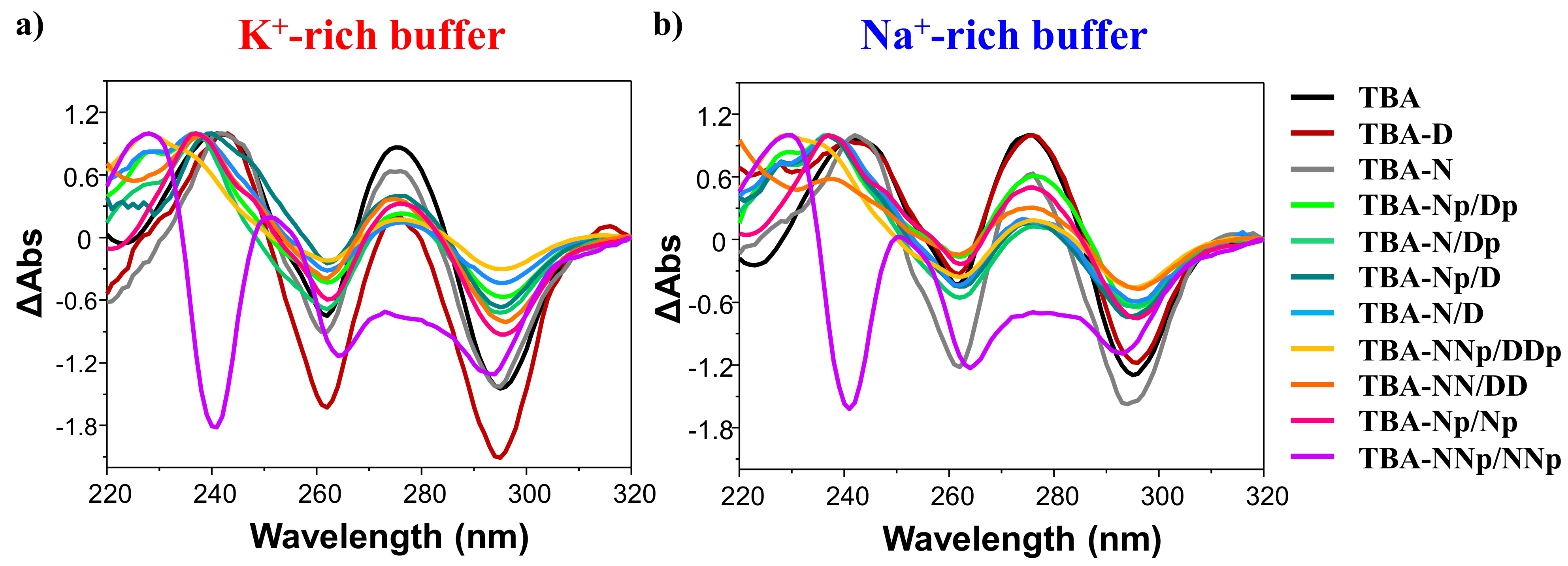
2.2.1.2. CD Spectroscopy Analysis: CD Spectra and CD Thermal Denaturation/Renaturation Measurements
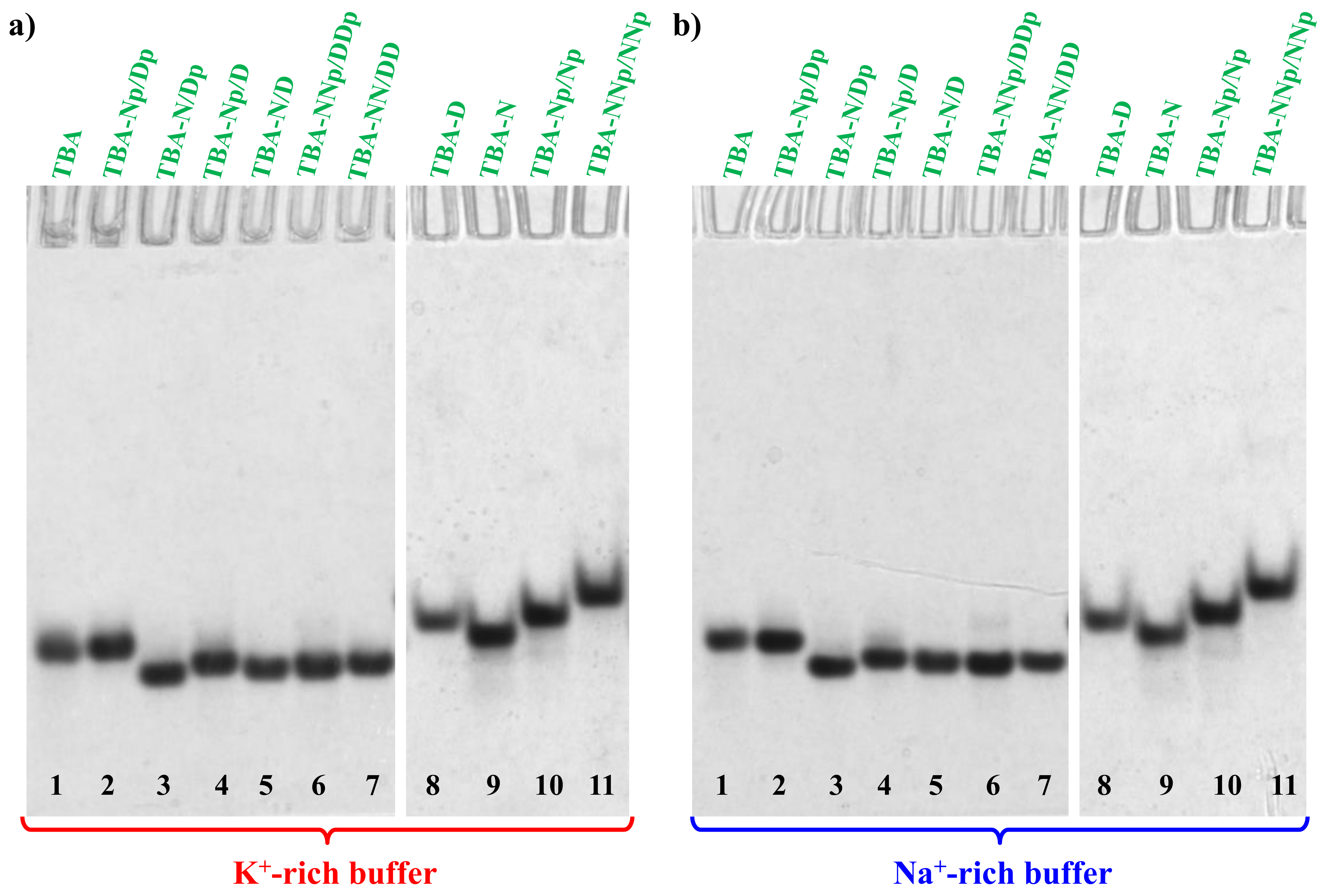
2.3. Non-Denaturing Polyacrylamide Gel Electrophoresis
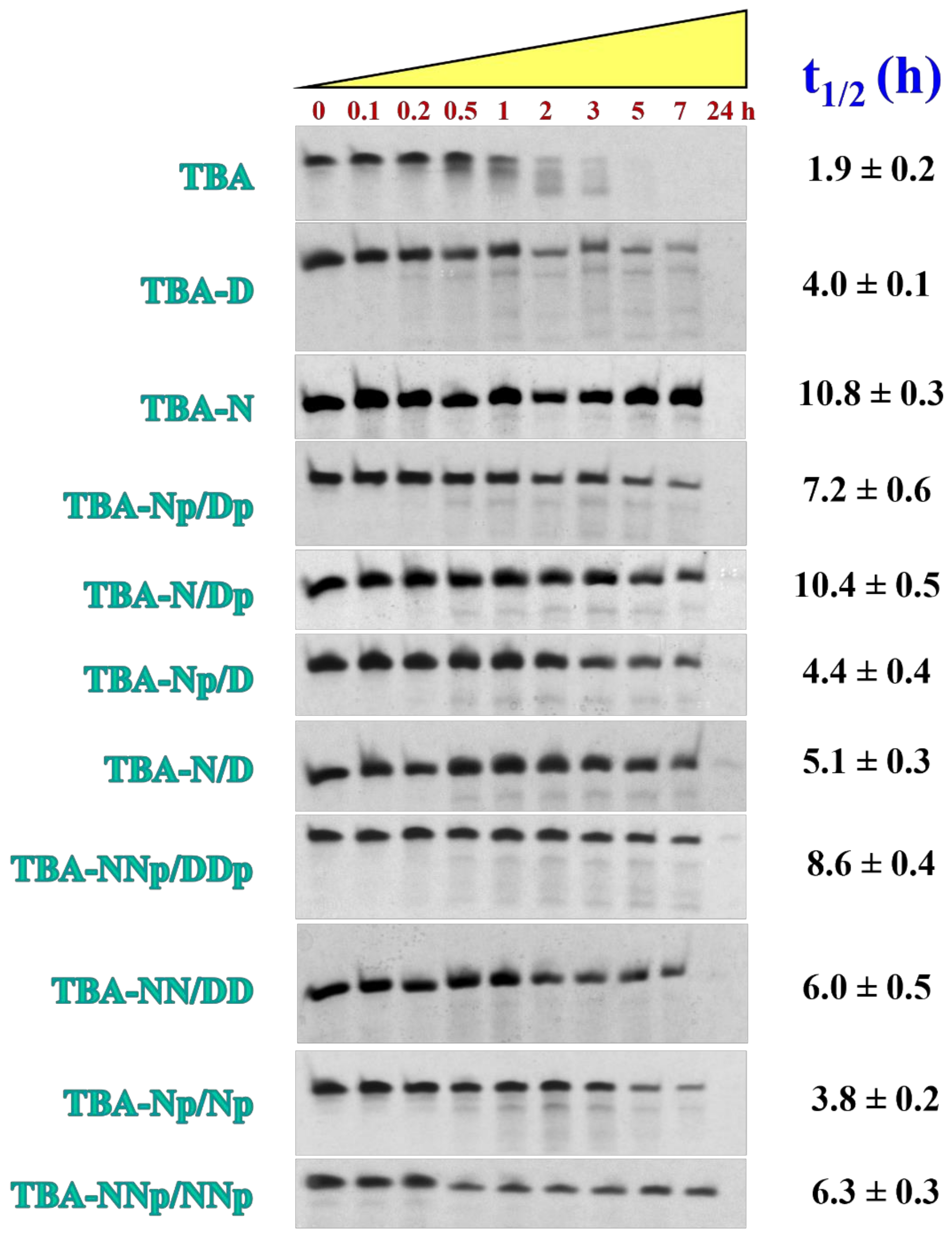
2.4. NDI and DAN Conjugating Groups Confer Higher Nucleases Stability to TBA
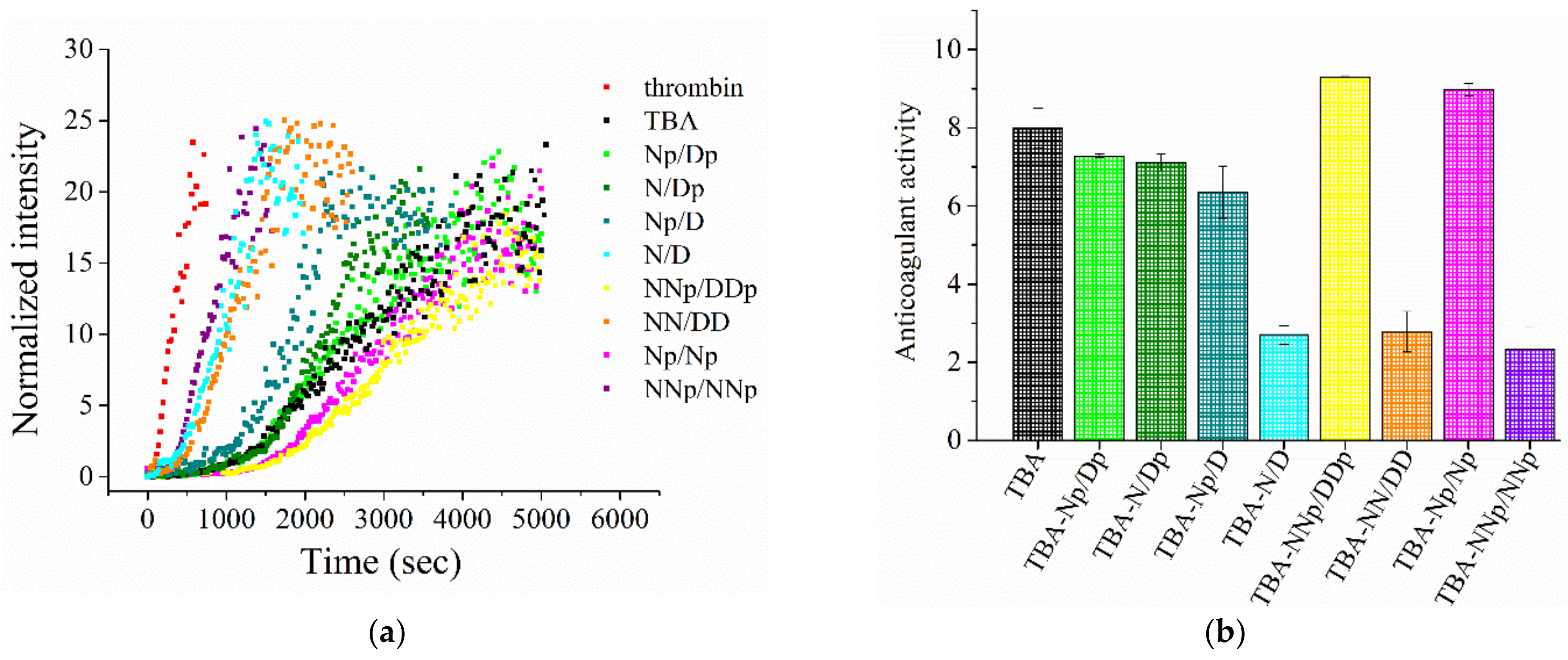
2.5. Coagulation Experiments
3. Materials and Methods
3.1. General Methods
3.2. Synthetic Procedures
3.3. Synthesis of DMTr-NDI-Q-Linker Solid Support 10
3.4. Synthesis of DMTr-DAN-Q-Linker Solid Support 13
3.5. Oligonucleotide Synthesis
3.6. Oligomer Treatment and Analysis
3.7. Preparation of the Oligonucleotide Samples
3.8. UV Spectroscopy
3.9. Polyacrylamide Gel Electrophoresis (PAGE) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [Green Version]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex forming aptamers—Characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [Green Version]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef]
- Antipova, O.M.; Zavyalova, E.G.; Golovin, A.V.; Pavlova, G.V.; Kopylov, A.M.; Reshetnikov, R.V. Advances in the application of modified nucleotides in SELEX technology. Biochemistry 2018, 83, 1161–1172. [Google Scholar] [CrossRef]
- Sola, M.; Menon, A.; Moreno, B.; Meraviglia-Crivelli, D.; Soldevilla, M.; Cartón-García, F.; Pastor, F. Aptamers against live targets: Is in vivo SELEX finally coming to edge? Mol. Ther. Nucleic Acids 2020, 21, 192–204. [Google Scholar] [CrossRef]
- Musumeci, D.; Platella, C.; Riccardi, C.; Moccia, F.; Montesarchio, D. Fluorescence sensing using DNA aptamers in cancer research and clinical diagnostics. Cancers 2017, 9, 174. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as therapeutics. Ann. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [Green Version]
- Ismail, S.I.; Alshaer, W. Therapeutic aptamers in discovery, preclinical and clinical stages. Adv. Drug Deliv. Rev. 2018, 134, 51–64. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Musumeci, D.; Montesarchio, D. Dimeric and multimeric DNA aptamers for highly effective protein recognition. Molecules 2020, 25, 5227. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Melone, M.A.B.; Montesarchio, D. Anti-VEGF DNA-based aptamers in cancer therapeutics and diagnostics. Med. Res. Rev. 2021, 41, 464–506. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Liu, B.; Lu, A.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef] [Green Version]
- Catuogno, S.; Esposito, C.L. Aptamer cell-based selection: Overview and advances. Biomedicines 2017, 5, 49. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the therapeutics and diagnostics pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef]
- Lapa, S.A.; Chudinov, A.V.; Timofeev, E.N. The toolbox for modified aptamers. Mol. Biotechnol. 2016, 58, 79–92. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diafa, S.; Hollenstein, M. Generation of aptamers with an expanded chemical repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [Green Version]
- Tolle, F.; Mayer, G. Dressed for success-applying chemistry to modulate aptamer functionality. Chem. Sci. 2013, 4, 60–67. [Google Scholar] [CrossRef]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [Green Version]
- Aviñó, A.; Fàbrega, C.; Tintoré, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef] [Green Version]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Montesarchio, D. G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects. Pharmacol. Ther. 2021, 217, 107649. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Giancola, C.; Randazzo, A.; Mazzarella, L.; Sica, F. Thrombin-aptamer recognition: A revealed ambiguity. Nucleic Acids Res. 2011, 39, 7858–7867. [Google Scholar] [CrossRef] [Green Version]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cera, E. Thrombin. Mol. Aspects Med. 2008, 29, 203–254. [Google Scholar] [CrossRef] [PubMed]
- Licari, L.G.; Kovacic, J.P. Thrombin physiology and pathophysiology. J. Vet. Emerg. Crit. Care 2009, 19, 11–22. [Google Scholar] [CrossRef]
- Zavyalova, E.G.; Ustinov, N.; Golovin, A.; Pavlova, G.; Kopylov, A. G-quadruplex aptamers to human thrombin versus other direct thrombin inhibitors: The focus on mechanism of action and drug efficiency as anticoagulants. Curr. Med. Chem. 2016, 23, 2230–2244. [Google Scholar] [CrossRef] [PubMed]
- Posma, J.J.N.; Posthuma, J.J.; Spronk, H.M.H. Coagulation and non-coagulation effects of thrombin. J. Thromb. Haemost. 2016, 14, 1908–1916. [Google Scholar] [CrossRef] [Green Version]
- Pica, A.; Russo Krauss, I.; Merlino, A.; Nagatoishi, S.; Sugimoto, N.; Sica, F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013, 280, 6581–6588. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Harrington, R.A.; Sullenger, B.A. The potential of aptamers as anticoagulants. Trends Cardiovasc. Med. 2005, 15, 41–45. [Google Scholar] [CrossRef]
- Schwienhorst, A. Direct thrombin inhibitors—A survey of recent developments. Cell Mol. Life Sci. 2006, 63, 2773–2791. [Google Scholar] [CrossRef]
- Mayer, G.; Rohrbach, F.; Pötzsch, B.; Müller, J. Aptamer-based modulation of blood coagulation. Hamostaseologie 2011, 31, 258–263. [Google Scholar] [CrossRef] [PubMed]
- De Napoli, L.; Messere, A.; Montesarchio, D.; Piccialli, G.; Santacroce, C.; Bonora, G.M. PEG-supported synthesis of cyclic oligodeoxyribonucleotides. Nucleosides Nucleotides 1993, 12, 21–30. [Google Scholar] [CrossRef]
- De Napoli, L.; Galeone, A.; Mayol, L.; Messere, A.; Montesarchio, D.; Piccialli, G. Automated solid phase synthesis of cyclic oligonucleotides: A further improvement. Bioorg. Med. Chem. 1995, 3, 1325–1329. [Google Scholar] [CrossRef]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Russo Krauss, I.; Paduano, L.; Oliva, R.; Petraccone, L.; Morvan, F.; Montesarchio, D. Stability is not everything: The case of the cyclization of the thrombin binding aptamer. ChemBioChem 2019, 20, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Russo Krauss, I.; Paduano, L.; Morvan, F.; Montesarchio, D. Fine-tuning the properties of the thrombin binding aptamer through cyclization: Effect of the 5′-3′ connecting linker on the aptamer stability and anticoagulant activity. Bioorg. Chem. 2020, 94, 103379. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Whyte, J.; Khuri, S.F.; Preiss, J.R.; Kurz, J.C.; Olson, K.; Hatala, P.; Boomer, R.M.; Fraone, J.M.; Brosnan, N.; Makim, A.; et al. Discovery of a potent, direct thrombin inhibiting aptamer. J. Thromb. Haemost. 2007, 5, P-S-067. [Google Scholar] [CrossRef]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Russo Krauss, I.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, 46, 12177–12185. [Google Scholar] [CrossRef] [Green Version]
- Riccardi, C.; Meyer, A.; Vasseur, J.J.; Cavasso, D.; Russo Krauss, I.; Paduano, L.; Morvan, F.; Montesarchio, D. Design, synthesis and characterization of cyclic NU172 analogues: A biophysical and biological insight. Int. J. Mol. Sci. 2020, 21, 3860. [Google Scholar] [CrossRef]
- Pérez de Carvasal, K.; Aissaoui, N.; Vergoten, G.; Bellot, G.; Vasseur, J.-J.; Smietana, M.; Morvan, F. Folding of phosphodiester-linked donor–acceptor oligomers into supramolecular nanotubes in water. Chem. Commun. 2021, 57, 4130–4133. [Google Scholar] [CrossRef]
- Scott Lokey, R.; Iverson, B.L. Synthetic molecules that fold into a pleated secondary structure in solution. Nature 1995, 375, 303–305. [Google Scholar] [CrossRef]
- Foster, R. Electron donor-acceptor complexes. J. Phys. Chem. 1980, 84, 2135–2141. [Google Scholar] [CrossRef]
- Cubberley, M.S.; Iverson, B.L. 1H NMR investigation of solvent effects in aromatic stacking interactions. J. Am. Chem. Soc. 2001, 123, 7560–7563. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, M.D.; Caruthers, M.H. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981, 103, 3185–3191. [Google Scholar] [CrossRef]
- Pon, R.T.; Yu, S. Hydroquinone-O,O’-diacetic acid (‘Q-linker’) as a replacement for succinyl and oxalyl linker arms in solid phase oligonucleotide synthesis. Nucleic Acids Res. 1997, 25, 3629–3635. [Google Scholar] [CrossRef] [Green Version]
- Zavyalova, E.G.; Tagiltsev, G.; Reshetnikov, R.; Arutyunyan, A.; Kopylov, A. Cation coordination alters the conformation of a thrombin-binding G-quadruplex DNA aptamer that affects inhibition of thrombin. Nucleic Acid Ther. 2016, 6, 299–308. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef] [Green Version]
- Largy, E.; Marchand, A.; Amrane, S.; Gabelica, V.; Mergny, J.L. Quadruplex turncoats: Cation-dependent folding and stability of quadruplex-DNA double switches. J. Am. Chem. Soc. 2016, 138, 2780–2792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mergny, J.-L.; Li, J.; Lacroix, L.; Amrane, S.; Chaires, J.B. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, e138. [Google Scholar] [CrossRef] [Green Version]
- Mergny, J.-L.; Lacroix, L. UV Melting of G-quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009, 37, 17.1.1–17.1.15. [Google Scholar] [CrossRef] [PubMed]
- Karsisiotis, A.I.; Hessari, N.M.A.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba da Silva, M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem.-Int. Ed. Eng. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Malgowska, M.; Gudanis, D.; Teubert, A.; Dominiak, G.; Gdaniec, Z. How to study G-quadruplex structures. J. Biotechnol. Comput. Biol. Bionanotechnol. 2012, 93, 381–390. [Google Scholar] [CrossRef]
- Dapić, V.; Abdomerović, V.; Marrington, R.; Peberdy, J.; Rodger, A.; Trent, J.O.; Bates, P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef] [Green Version]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [Green Version]
- Masiero, S.; Trotta, R.; Pieraccini, S.; De Tito, S.; Perone, R.; Randazzo, A.; Spada, G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010, 8, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.; Spada, G.P.; Webba da Silva, M. Circular dichroism of quadruplex structures. Top Curr. Chem. 2013, 330, 67–86. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of quadruplex DNA structure by circular dichroism. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 17.8.1–17.8.16. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem.-Int. Ed. Eng. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Poniková, S.; Antalík, M.; Hianik, T. A circular dichroism study of the stability of guanine quadruplexes of thrombin DNA aptamers at presence of K+ and Na+ ions. Gen. Physiol. Biophys. 2008, 27, 271–277. [Google Scholar] [PubMed]
- Saintomé, C.; Amrane, S.; Mergny, J.L.; Alberti, P. The exception that confirms the rule: A higher-order telomeric G-quadruplex structure more stable in sodium than in potassium. Nucleic Acids Res. 2016, 44, 2926–2935. [Google Scholar] [CrossRef]
- Platella, C.; Napolitano, E.; Riccardi, C.; Musumeci, D.; Montesarchio, D. Disentangling the structure-activity relationships of naphthalene diimides as anticancer G-quadruplex-targeting drugs. J. Med. Chem. 2021, 64, 3578–3603. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, M.; Kaluzhny, D.; Shchyolkina, A.; Borisova, O.; Smirnov, I.; Pozmogova, G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys. Chem. 2010, 146, 1–6. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Napolitano, V.; Petraccone, L.; Troisi, R.; Spiridonova, V.; Mattia, C.A.; Sica, F. Duplex/quadruplex oligonucleotides: Role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018, 107, 1697–1705. [Google Scholar] [CrossRef]
- Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative mechanisms underlying high inhibitory activities of bimodular DNA aptamers to thrombin. Biomolecules 2019, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Marky, L.A.; Breslauer, K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 1987, 26, 1601–1620. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Martino, L.; Virno, A.; Randazzo, A.; Virgilio, A.; Esposito, V.; Giancola, C.; Bucci, M.; Cirino, G.; Mayol, L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006, 34, 6653–6662. [Google Scholar] [CrossRef] [PubMed]
- Virno, A.; Randazzo, A.; Giancola, C.; Bucci, M.; Cirino, G.; Mayol, L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg. Med. Chem. 2007, 15, 5710–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virgilio, A.; Petraccone, L.; Vellecco, V.; Bucci, M.; Varra, M.; Irace, C.; Santamaria, R.; Pepe, A.; Mayol, L.; Esposito, V.; et al. Site-specific replacement of the thymine methyl group by fluorine in thrombin binding aptamer significantly improves structural stability and anticoagulant activity. Nucleic Acids Res. 2015, 43, 10602–10611. [Google Scholar] [CrossRef] [Green Version]
- Petraccone, L.; Erra, E.; Nasti, L.; Galeone, A.; Randazzo, A.; Mayol, L.; Barone, G.; Giancola, C. Effect of a modified thymine on the structure and stability of [d(TGGGT)]4 quadruplex. Int. J. Biol. Macromol. 2003, 31, 131–137. [Google Scholar] [CrossRef]
- Petraccone, L.; Erra, E.; Esposito, V.; Randazzo, A.; Mayol, L.; Nasti, L.; Barone, G.; Giancola, C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry 2004, 43, 4877–4884. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M.; Gmeiner, W.H.; Marky, L.A. Unfolding of G-quadruplexes: Energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B 2006, 110, 6962–6969. [Google Scholar] [CrossRef] [PubMed]
- Petraccone, L.; Spink, C.; Trent, J.O.; Garbett, N.C.; Mekmaysy, C.S.; Giancola, C.; Chaires, J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011, 133, 20951–20961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the conformational behaviour and aggregation properties of lipid-conjugated AS1411 aptamers. Int. J. Biol. Macromol. 2018, 118, 1384–1399. [Google Scholar] [CrossRef]
- Moccia, F.; Riccardi, C.; Musumeci, D.; Leone, S.; Oliva, R.; Petraccone, L.; Montesarchio, D. Insights into the G-rich VEGF-binding aptamer V7t1: When two G-quadruplexes are better than one! Nucleic Acids Res. 2019, 47, 8318–8331. [Google Scholar] [CrossRef] [Green Version]
- Dougan, H.; Lyster, D.M.; Vo, C.V.; Stafford, A.; Weitz, J.I.; Hobbs, J.B. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Biol. 2000, 27, 289–297. [Google Scholar] [CrossRef]
- Uehara, S.; Shimada, N.; Takeda, Y.; Koyama, Y.; Takei, Y.; Ando, H.; Satoh, S.; Uno, A.; Sakurai, K. 3′ poly(dA)-tailed thrombin DNA aptamer to increase DNase-resistance and clotting inhibitory activity. Bull. Chem. Soc. Jpn. 2008, 81, 1485–1491. [Google Scholar] [CrossRef]
- Kasahara, Y.; Kitadume, S.; Morihiro, K.; Kuwahara, M.; Ozaki, H.; Sawai, H.; Imanishi, T.; Obika, S. Effect of 3′-end capping of aptamer with various 2′,4′-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorg. Med. Chem. 2010, 20, 1626–1629. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Russo Krauss, I.; Musumeci, D.; Morvan, F.; Meyer, A.; Vasseur, J.J.; Paduano, L.; Montesarchio, D. Fluorescent thrombin binding aptamer-tagged nanoparticles for an efficient and reversible control of thrombin activity. ACS Appl. Mater. Interfaces 2017, 9, 35574–35587. [Google Scholar] [CrossRef]
- Hoke, G.D.; Draper, K.; Freier, S.M.; Gonzalez, C.; Driver, V.B.; Zounes, M.C.; Ecker, D.J. Effects of phosphorothioate capping on antisense oligonucleotide stability, hybridization and antiviral efficacy versus herpes simplex virus infection. Nucleic Acids Res. 1991, 19, 5743–5748. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Petraccone, L.; Pagano, B.; Giancola, C. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods 2012, 57, 76–83. [Google Scholar] [CrossRef]
- Petersheim, M.; Turner, D.H. Base-stacking and base-pairing contributions to helix stability: Thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry 1983, 22, 256–263. [Google Scholar] [CrossRef]
- Riccardi, C.; Musumeci, D.; Platella, C.; Gaglione, R.; Arciello, A.; Montesarchio, D. Tuning the polymorphism of the anti-VEGF G-rich V7t1 aptamer by covalent dimeric constructs. Int. J. Mol. Sci. 2020, 21, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
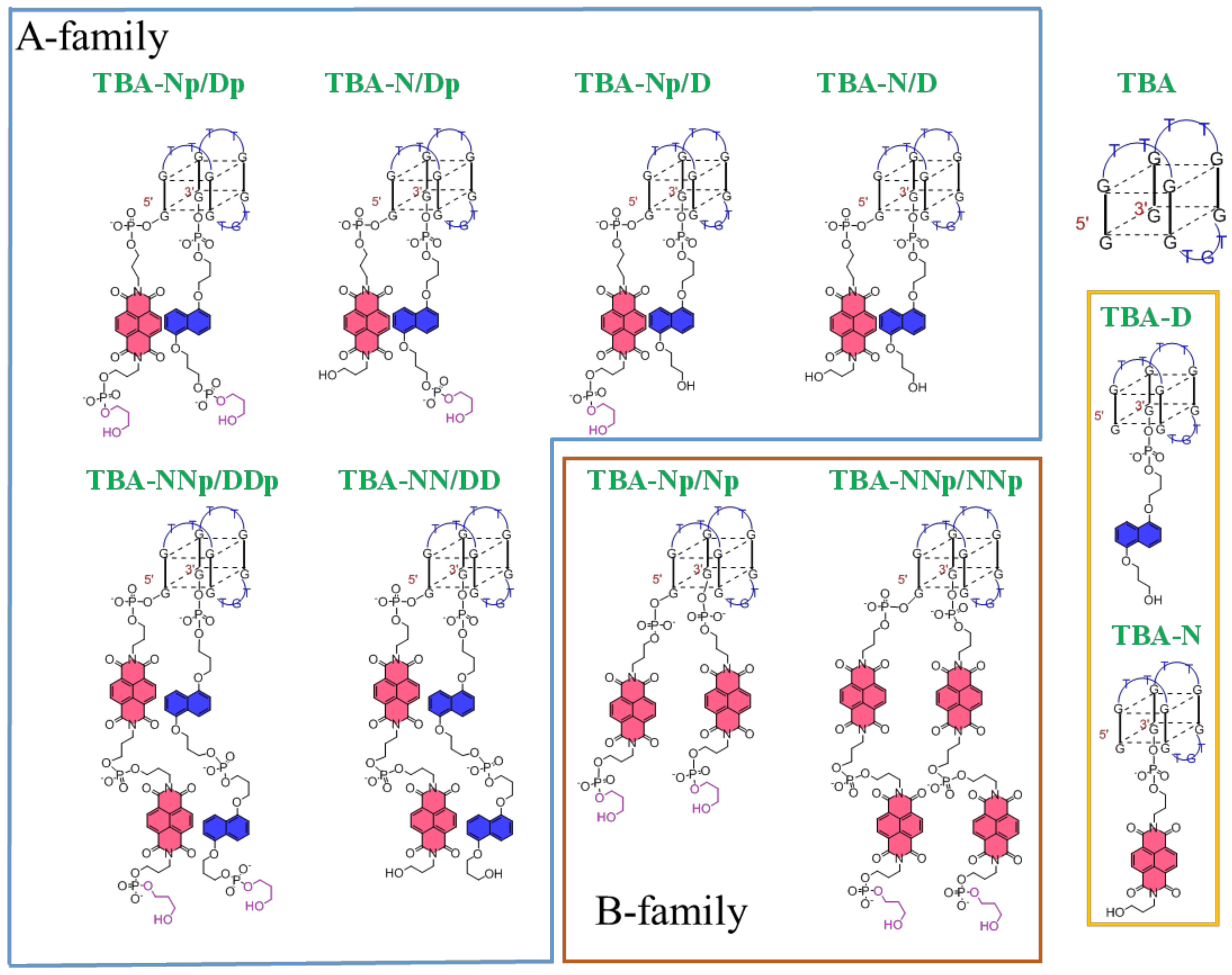


| Aptamer | CD-Melting and Annealing Experiments | |||||
|---|---|---|---|---|---|---|
| K+-Rich Buffer | Na+-Rich Buffer | |||||
| Melting Tm (°C) | ΔTm (°C) | Annealing Tm (°C) | Melting Tm (°C) | ΔTm (°C) | Annealing Tm (°C) | |
| TBA | 51 | 0 | 49 | 36 | - | 35 |
| TBA-D | 45 | −6 | 43 | 31 | −5 | 30 |
| TBA-N | 64 | +13 | 62 | 51 | +15 | 49 |
| TBA-Np/Dp | 51 | 0 | 51 | 38 | +2 | 37 |
| TBA-N/Dp | 62 | +11 | 61 | 48 | +12 | 48 |
| TBA-Np/D | 57 | +6 | 56 | 42 | +6 | 42 |
| TBA-N/D | 68 | +17 | 66 | 51 | +15 | 49 |
| TBA-NNp/DDp | 62 | +11 | 61 | 44 | +8 | 43 |
| TBA-NN/DD | 61 | +10 | 60 | 42 | +6 | 40 |
| TBA-Np/Np | 49 | −2 | 48 | 34 | −2 | 33 |
| TBA-NNp/NNp * | 56 | +5 | 55 | 44 | +8 | 43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez de Carvasal, K.; Riccardi, C.; Russo Krauss, I.; Cavasso, D.; Vasseur, J.-J.; Smietana, M.; Morvan, F.; Montesarchio, D. Charge-Transfer Interactions Stabilize G-Quadruplex-Forming Thrombin Binding Aptamers and Can Improve Their Anticoagulant Activity. Int. J. Mol. Sci. 2021, 22, 9510. https://doi.org/10.3390/ijms22179510
Pérez de Carvasal K, Riccardi C, Russo Krauss I, Cavasso D, Vasseur J-J, Smietana M, Morvan F, Montesarchio D. Charge-Transfer Interactions Stabilize G-Quadruplex-Forming Thrombin Binding Aptamers and Can Improve Their Anticoagulant Activity. International Journal of Molecular Sciences. 2021; 22(17):9510. https://doi.org/10.3390/ijms22179510
Chicago/Turabian StylePérez de Carvasal, Kévan, Claudia Riccardi, Irene Russo Krauss, Domenico Cavasso, Jean-Jacques Vasseur, Michael Smietana, François Morvan, and Daniela Montesarchio. 2021. "Charge-Transfer Interactions Stabilize G-Quadruplex-Forming Thrombin Binding Aptamers and Can Improve Their Anticoagulant Activity" International Journal of Molecular Sciences 22, no. 17: 9510. https://doi.org/10.3390/ijms22179510







