Novel Lytic Enzyme of Prophage Origin from Clostridium botulinum E3 Strain Alaska E43 with Bactericidal Activity against Clostridial Cells
Abstract
:1. Introduction
2. Results
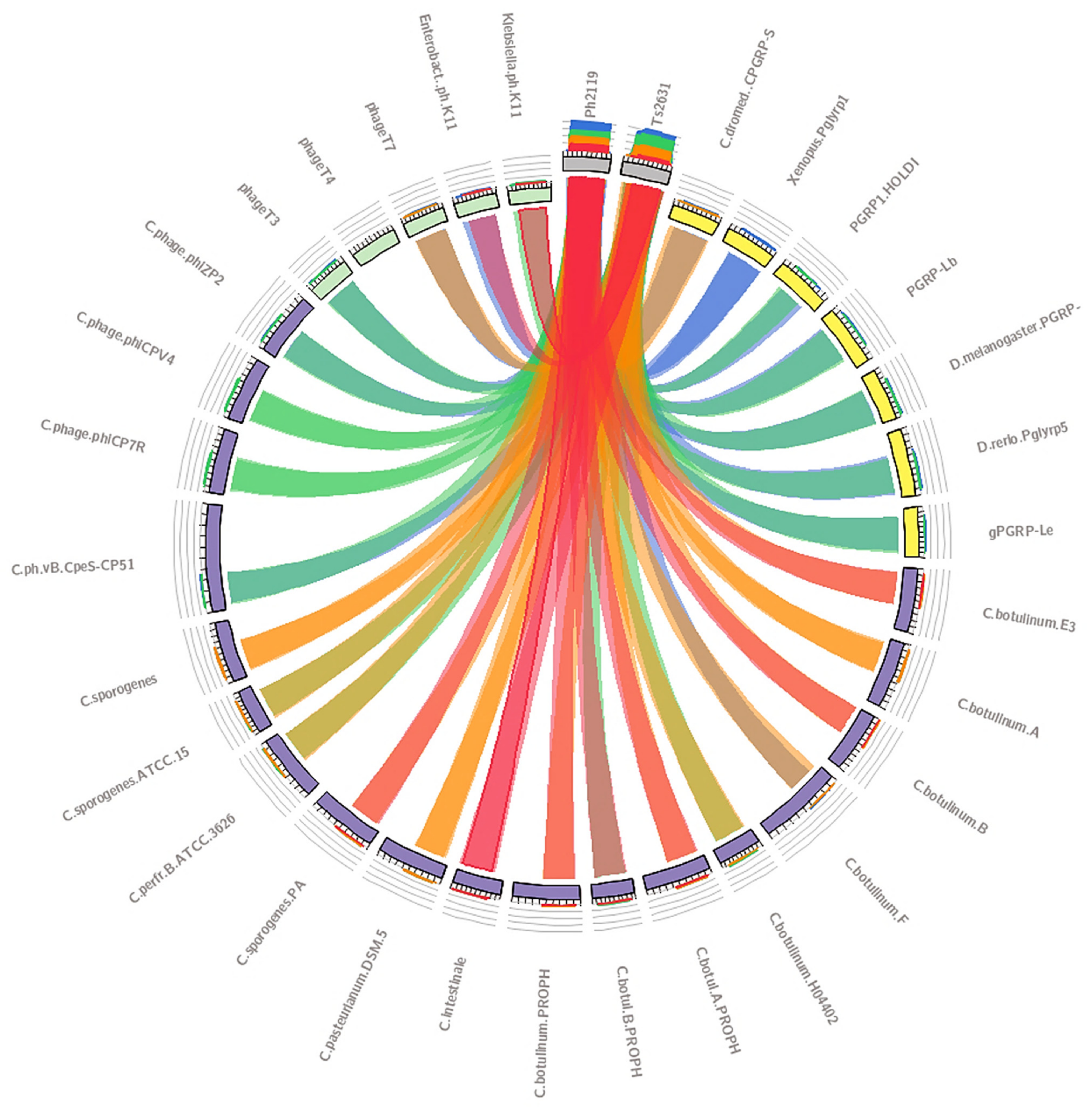
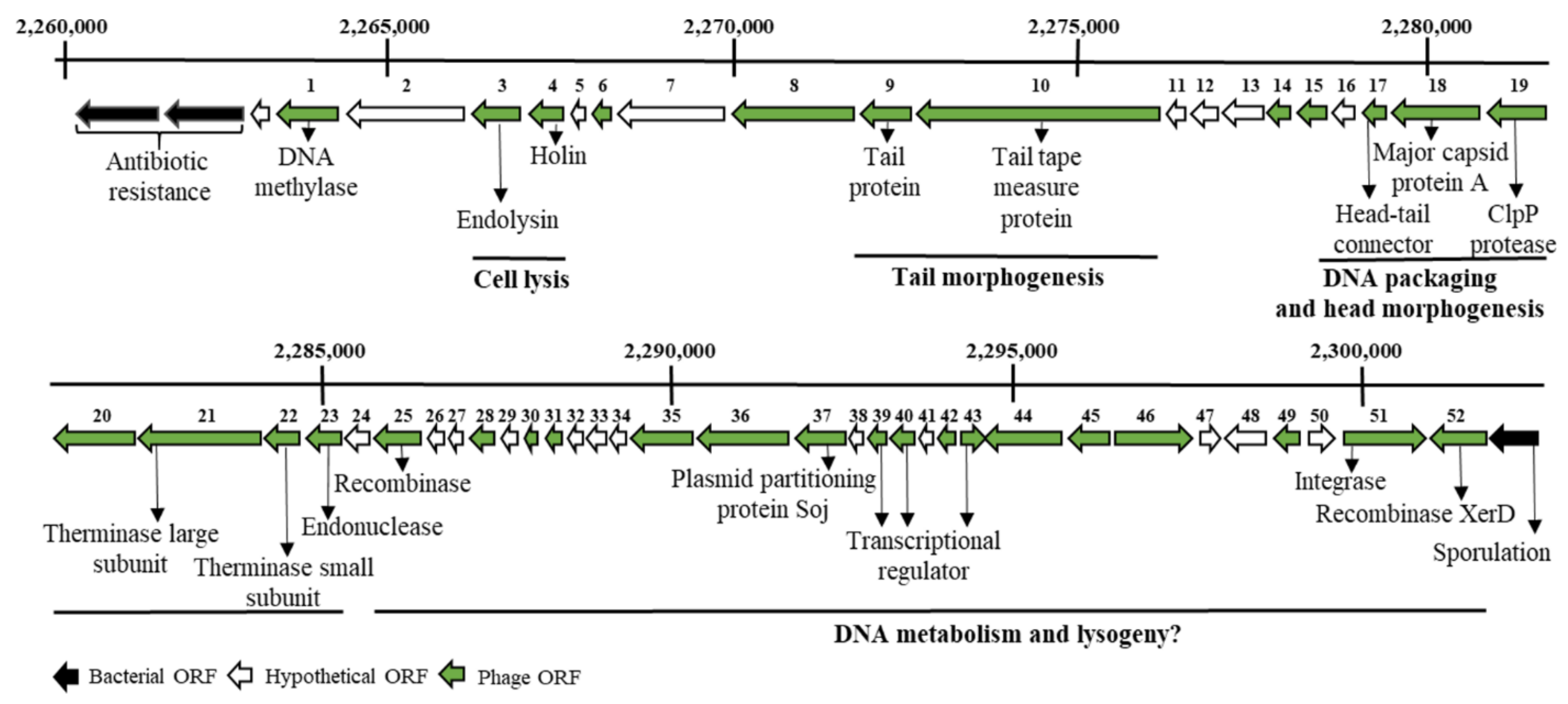
2.1. In Silico Analysis in Search of Lytic Enzymes
2.2. Expression and Purification of LysB Endolysin
2.3. Bacteriolytic Spectrum of LysB Endolysin
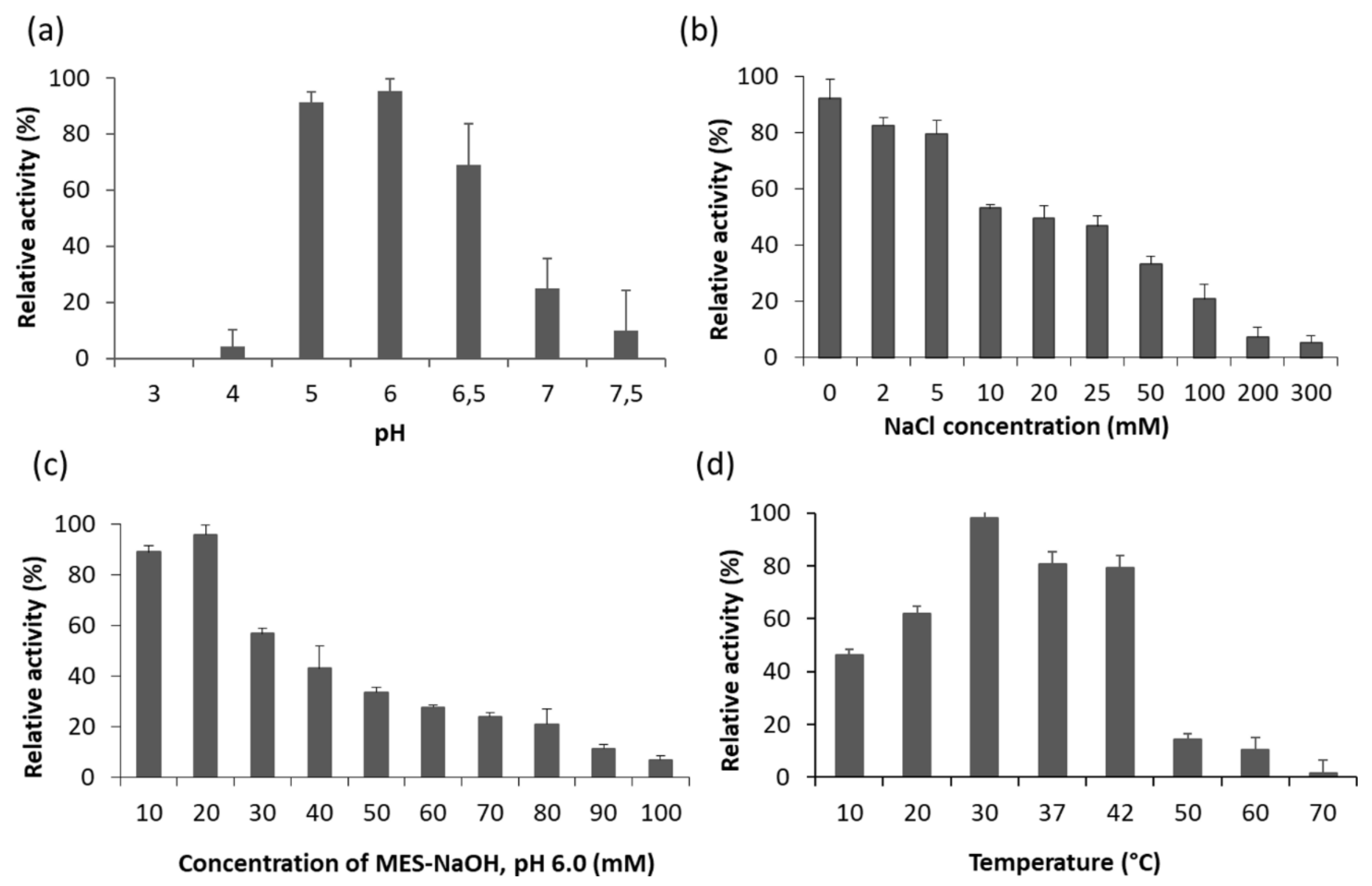
2.4. Optimal Conditions for LysB activity
2.5. Electron Microscopy Experiments
2.6. Oligomeric State of LysB Endolysin
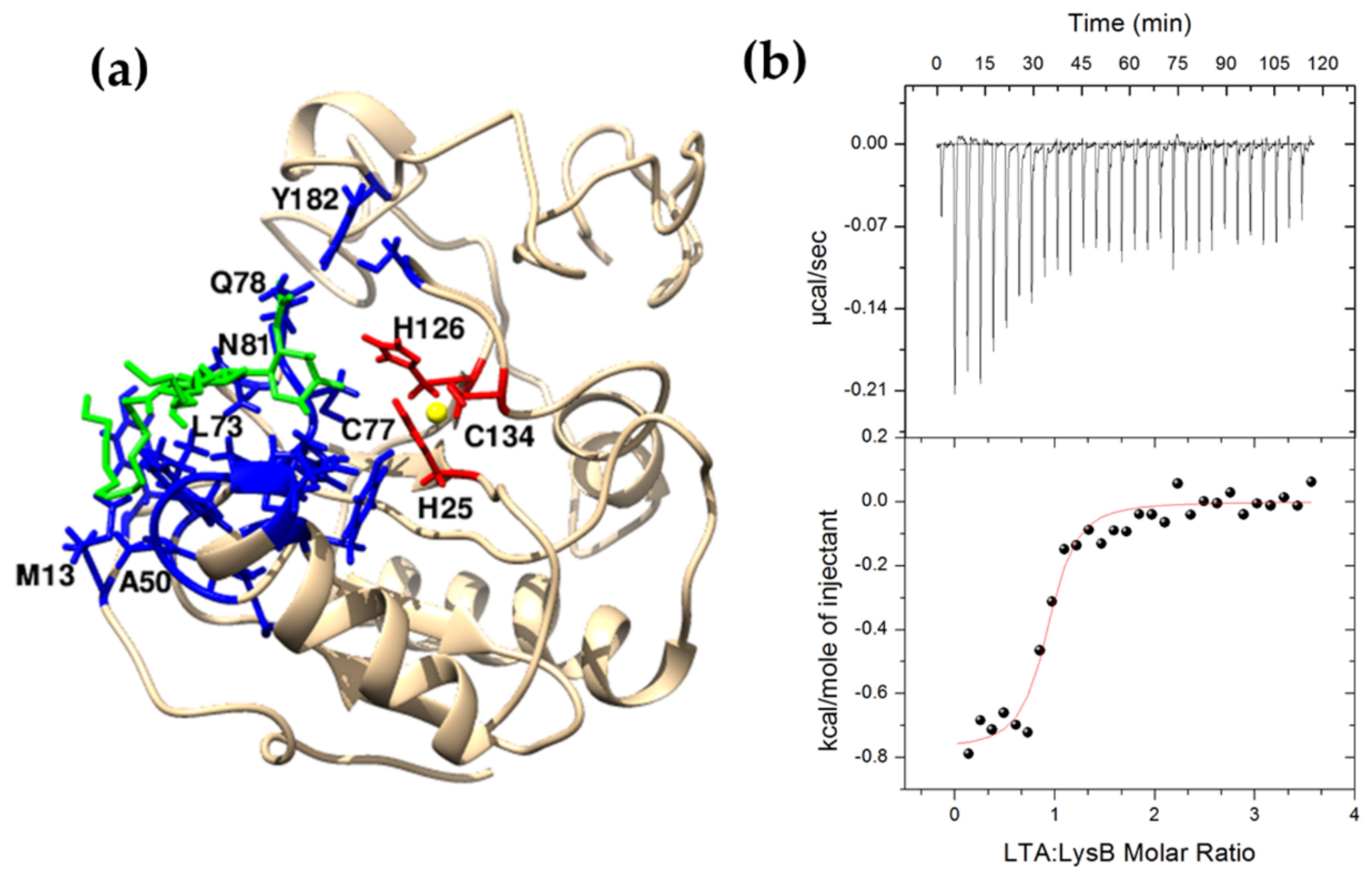
2.7. Interaction between LysB and Lipoteichoic Acid
2.8. Functional Analysis of the LysB Catalytic Site
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Computational Analysis and Molecular Modelling
4.3. DNA Manipulations
4.4. Expression and Purification of LysB Endolysin and Its Substitution Variants
4.5. Isothermal Titration Calorimetry
4.6. Testing the LysB Endolysin Optimum
4.7. Antibacterial Spectrum of LysB Endolysin
4.8. Size-Exclusion Chromatography (SEC) and Analytical Ultracentrifugation (AUC)
4.9. Transmission Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shukla, H.D.; Sharma, S.K. Clostridium botulinum: A bug with beauty and weapon. Crit. Rev. Microbiol. 2005, 31, 11–18. [Google Scholar] [CrossRef]
- Poulain, B.; Popoff, M.R. Why Are Botulinum Neurotoxin-Producing Bacteria So Diverse and Botulinum Neurotoxins So Toxic? Toxins 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Sobel, J. Botulism. Clin. Infect. Dis. 2005, 41, 1167–1173. [Google Scholar] [CrossRef]
- Mazuet, C.; Legeay, C.; Sautereau, J.; Ma, L.; Bouchier, C.; Bouvet, P.; Popoff, M.R. Diversity of Group I and II Clostridium botulinum Strains from France Including Recently Identified Subtypes. Genome Biol. Evol. 2016, 8, 1643–1660. [Google Scholar] [CrossRef]
- Lonati, D.; Schicchi, A.; Crevani, M.; Buscaglia, E.; Scaravaggi, G.; Maida, F.; Cirronis, M.; Petrolini, V.M.; Locatelli, C.A. Foodborne Botulism: Clinical Diagnosis and Medical Treatment. Toxins 2020, 12, 509. [Google Scholar] [CrossRef]
- Carter, A.T.; Peck, M.W. Genomes, neurotoxins and biology of Clostridium botulinum Group I and Group II. Res. Microbiol. 2015, 166, 303–317. [Google Scholar] [CrossRef] [Green Version]
- Brunt, J.; van Vliet, A.H.M.; Stringer, S.C.; Carter, A.T.; Lindström, M.; Peck, M.W. Pan-Genomic Analysis of Clostridium botulinum Group II (Non-Proteolytic, C. botulinum) Associated with Foodborne Botulism and Isolated from the Environment. Toxins 2020, 12, 306. [Google Scholar] [CrossRef]
- Skarin, H.; Håfström, T.; Westerberg, J.; Segerman, B. Clostridium botulinum group III: A group with dual identity shaped by plasmids, phages and mobile elements. BMC Genom. 2011, 12, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, S.I.; Zhang, J.; Zhang, S.; Shoemaker, C.B.; Dong, M. Delivery of single-domain antibodies into neurons using a chimeric toxin-based platform is therapeutic in mouse models of botulism. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Servick, K. Decoy toxin harnessed to fight botulism. Science 2021, 371, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Kwon, J.G.; O’Sullivan, D.J.; Ryu, S.; Lee, J.H. Development of an endolysin enzyme and its cell wall-binding domain protein and their applications for biocontrol and rapid detection of Clostridium perfringens in food. Food Chem. 2021, 345, 128562. [Google Scholar] [CrossRef]
- Hammond, R.W.; Swift, S.M.; Foster-Frey, J.A.; Kovalskaya, N.Y.; Donovan, D.M. Optimized production of a biologically active Clostridium perfringens glycosyl hydrolase phage endolysin PlyCP41 in plants using virus-based systemic expression. BMC Biotechnol. 2019, 19, 101. [Google Scholar] [CrossRef]
- Zimmer, M.; Vukov, N.; Scherer, S.; Loessner, M.J. The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 2002, 68, 5311–5317. [Google Scholar] [CrossRef] [Green Version]
- Simmons, M.; Donovan, D.M.; Siragusa, G.R.; Seal, B.S. Recombinant expression of two bacteriophage proteins that lyse Clostridium perfringens and share identical sequences in the C-terminal cell wall binding domain of the molecules but are dissimilar in their N-terminal active domains. J. Agric. Food Chem. 2010, 58, 10330–10337. [Google Scholar] [CrossRef] [Green Version]
- Nariya, H.; Miyata, S.; Tamai, E.; Sekiya, H.; Maki, J.; Okabe, A. Identification and characterization of a putative endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 90, 1973–1979. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Ossiprandi, M.C.; Rumah, K.R.; Fischetti, V.A. Lytic enzyme discovery through multigenomic sequence analysis in Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 89, 1783–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gervasi, T.; Horn, N.; Wegmann, U.; Dugo, G.; Narbad, A.; Mayer, M.J. Expression and delivery of an endolysin to combat Clostridium perfringens. Appl. Microbiol. Biotechnol. 2014, 98, 2495–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens Virulent Bacteriophage CPS2 and Its Thermostable Endolysin LysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritter, S.C.; Hackel, B.J. Validation and Stabilization of a Prophage Lysin of Clostridium perfringens by Using Yeast Surface Display and Coevolutionary Models. Appl. Environ. Microbiol. 2019, 85, e00054-19. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.I.; Draper, L.A.; Ross, R.P.; Hill, C. Bacteriophage endolysins as a potential weapon to combat Clostridioides difficile infection. Gut Microbes 2020, 12, 1813533. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Euler, C.W.; Delaune, A.; Fischetti, V.A. Using a Novel Lysin To Help Control Clostridium difficile Infections. Antimicrob. Agents Chemother. 2015, 59, 7447–7457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, M.J.; Narbad, A.; Gasson, M.J. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 2008, 190, 6734–6740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, M.J.; Gasson, M.J.; Narbad, A. Genomic sequence of bacteriophage ATCC 8074-B1 and activity of its endolysin and engineered variants against Clostridium sporogenes. Appl. Environ. Microbiol. 2012, 78, 3685–3692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, M.J.; Payne, J.; Gasson, M.J.; Narbad, A. Genomic sequence and characterization of the virulent bacteriophage phiCTP1 from Clostridium tyrobutyricum and heterologous expression of its endolysin. Appl. Environ. Microbiol. 2010, 76, 5415–5422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Lahti, M.; Douillard, F.P.; Korkeala, H.; Lindström, M. Phage lysin that specifically eliminates Clostridium botulinum Group I cells. Sci. Rep. 2020, 10, 21571. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.J.; Garefalaki, V.; Spoerl, R.; Narbad, A.; Meijers, R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J. Bacteriol. 2011, 193, 5477–5486. [Google Scholar] [CrossRef] [Green Version]
- Dunne, M.; Leicht, S.; Krichel, B.; Mertens, H.D.; Thompson, A.; Krijgsveld, J.; Svergun, D.I.; Gómez-Torres, N.; Garde, S.; Uetrecht, C.; et al. Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product. J. Biol. Chem. 2016, 291, 4882–4893. [Google Scholar] [CrossRef] [Green Version]
- Plotka, M.; Kaczorowska, A.K.; Stefanska, A.; Morzywolek, A.; Fridjonsson, O.H.; Dunin-Horkawicz, S.; Kozlowski, L.; Hreggvidsson, G.O.; Kristjansson, J.K.; Dabrowski, S.; et al. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 2014, 80, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Plotka, M.; Kaczorowska, A.K.; Morzywolek, A.; Makowska, J.; Kozlowski, L.P.; Thorisdottir, A.; Skírnisdottir, S.; Hjörleifsdottir, S.; Fridjonsson, O.H.; Hreggvidsson, G.O.; et al. Biochemical Characterization and Validation of a Catalytic Site of a Highly Thermostable Ts2631 Endolysin from the Thermus scotoductus Phage vB_Tsc2631. PLoS ONE 2015, 10, e0137374. [Google Scholar] [CrossRef] [Green Version]
- Aevarsson, A.; Kaczorowska, A.K.; Adalsteinsson, B.T.; Ahlqvist, J.; Al-Karadaghi, S.; Altenbuchner, J.; Arsin, H.; Átlasson, Ú.; Brandt, D.; Cichowicz-Cieślak, M.; et al. Going to extremes—a metagenomic journey into the dark matter of life. FEMS Microbiol. Lett. 2021, 368, fnab067. [Google Scholar] [CrossRef]
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.K.; Kaczorowski, T. Ts2631 Endolysin from the Extremophilic Thermus scotoductus Bacteriophage vB_Tsc2631 as an Antimicrobial Agent against Gram-Negative Multidrug-Resistant Bacteria. Viruses 2019, 11, 657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, T.; Haque, S.; Somvanshi, P. Comparative assessment of the therapeutic drug targets of C. botulinum ATCC 3502 and C. difficile str. 630 using in silico subtractive proteomics approach. J. Cell. Biochem. 2019, 120, 16160–16184. [Google Scholar] [CrossRef]
- Weigand, M.R.; Pena-Gonzalez, A.; Shirey, T.B.; Broeker, R.G.; Ishaq, M.K.; Konstantinidis, K.T.; Raphael, B.H. Implications of Genome-Based Discrimination between Clostridium botulinum Group I and Clostridium sporogenes Strains for Bacterial Taxonomy. Appl. Environ. Microbiol. 2015, 81, 5420–5429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brüssow, H.; Desiere, F. Comparative phage genomics and the evolution of Siphoviridae: Insights from dairy phages. Mol. Microbiol. 2001, 39, 213–222. [Google Scholar] [CrossRef]
- Proux, C.; van Sinderen, D.; Suarez, J.; Garcia, P.; Ladero, V.; Fitzgerald, G.F.; Desiere, F.; Brüssow, H. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 2002, 184, 6026–6036. [Google Scholar] [CrossRef] [Green Version]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, X.; Pflugrath, J.W.; Studier, F.W. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 1994, 91, 4034–4038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziarski, R.; Kashyap, D.R.; Gupta, D. Mammalian peptidoglycan recognition proteins kill bacteria by activating two-component systems and modulate microbiome and inflammation. Microb. Drug Resist. 2012, 18, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Dube, D.; Singh, A.; Mishra, B.; Singh, N.; Sinha, M.; Dey, S.; Kaur, P.; Mitra, D.K.; Sharma, S.; et al. Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein. J. Biol. Chem. 2011, 286, 16208–16217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eugster, M.R.; Haug, M.C.; Huwiler, S.G.; Loessner, M.J. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol. Microbiol. 2011, 81, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Byun, M.; Oh, B.H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 2003, 4, 787–793. [Google Scholar] [CrossRef]
- Ugorcakova, J.; Bukovska, G. Lysins and Holins: Tools of Phage-Induced Lysis. Biologia 2003, 58, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Volozhantsev, N.V.; Oakley, B.B.; Morales, C.A.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Popova, A.V.; Zhilenkov, E.L.; Garrish, J.K.; Schegg, K.M.; et al. Molecular characterization of podoviral bacteriophages virulent for Clostridium perfringens and their comparison with members of the Picovirinae. PLoS ONE 2012, 7, e38283. [Google Scholar] [CrossRef] [Green Version]
- Mehta, K.K.; Paskaleva, E.E.; Wu, X.; Grover, N.; Mundra, R.V.; Chen, K.; Zhang, Y.; Yang, Z.; Feng, H.; Dordick, J.S.; et al. Newly identified bacteriolytic enzymes that target a wide range of clinical isolates of Clostridium difficile. Biotechnol. Bioeng. 2016, 113, 2568–2576. [Google Scholar] [CrossRef] [Green Version]
- Peck, M.W.; Stringer, S.C. The safety of pasteurised in-pack chilled meat products with respect to the foodborne botulism hazard. Meat Sci. 2005, 70, 461–475. [Google Scholar] [CrossRef]
- Swift, S.M.; Waters, J.J.; Rowley, D.T.; Oakley, B.B.; Donovan, D.M. Characterization of two glycosyl hydrolases, putative prophage endolysins, that target Clostridium perfringens. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Seal, B.S.; Garrish, J.K.; Oakley, B.B.; Hiett, K.; Yeh, H.Y.; Woolsey, R.; Schegg, K.M.; Line, J.E.; Donovan, D.M. A Thermophilic Phage Endolysin Fusion to a Clostridium perfringens-Specific Cell Wall Binding Domain Creates an Anti-Clostridium Antimicrobial with Improved Thermostability. Viruses 2015, 7, 3019–3034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briers, Y.; Lavigne, R. Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol. 2015, 10, 377–390. [Google Scholar] [CrossRef]
- Plotka, M.; Sancho-Vaello, E.; Dorawa, S.; Kaczorowska, A.K.; Kozlowski, L.P.; Kaczorowski, T.; Zeth, K. Structure and function of the Ts2631 endolysin of Thermus scotoductus phage vB_Tsc2631 with unique N-terminal extension used for peptidoglycan binding. Sci. Rep. 2019, 9, 1261. [Google Scholar] [CrossRef] [Green Version]
- Monterroso, B.; Sáiz, J.L.; García, P.; García, J.L.; Menéndez, M. Insights into the structure-function relationships of pneumococcal cell wall lysozymes, LytC and Cpl-1. J. Biol. Chem. 2008, 283, 28618–28628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höltje, J.V.; Tomasz, A. Lipoteichoic acid: A specific inhibitor of autolysin activity in Pneumococcus. Proc. Natl. Acad. Sci. USA 1975, 72, 1690–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular cloning: A laboratory manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, L.P. IPC—Isoelectric Point Calculator. Biol. Direct 2016, 11, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Shuvo, M.H.; Gulfam, M.; Bhattacharya, D. DeepRefiner: High-accuracy protein structure refinement by deep network calibration. Nucleic Acids Res. 2021, 49, W147–W152. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panuszko, A.; Bruździak, P.; Zielkiewicz, J.; Wyrzykowski, D.; Stangret, J. Effects of urea and trimethylamine-N-oxide on the properties of water and the secondary structure of hen egg white lysozyme. J. Phys. Chem. B 2009, 113, 14797–14809. [Google Scholar] [CrossRef] [PubMed]
- Plotka, M.; Szadkowska, M.; Håkansson, M.; Kovačič, R.; Al-Karadaghi, S.; Walse, B.; Werbowy, O.; Kaczorowska, A.K.; Kaczorowski, T. Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N-Terminal Extension. Int. J. Mol. Sci. 2020, 21, 4894. [Google Scholar] [CrossRef]
- Donovan, D.M.; Dong, S.; Garrett, W.; Rousseau, G.M.; Moineau, S.; Pritchard, D.G. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 2006, 72, 2988–2996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, P.H.; Schuck, P. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput. Phys. Commun. 2008, 178, 105–120. [Google Scholar] [CrossRef] [Green Version]


| Variants | Relative Activity (%) |
|---|---|
| LysB | 100.0 ± 4.4 |
| LysB + 5 mM EDTA a | 0.0 |
| LysB + 0.1 mM Zn2+ b | 42.4 ± 1.8 |
| LysB + 1 mM Zn2+ c | 0.0 |
| H25N | Nd |
| Y54F | 42.0 ± 4.4 |
| H126N | 37.0 ± 2.9 |
| S132K | 0.0 |
| C134S | 55.0 ± 9.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morzywolek, A.; Plotka, M.; Kaczorowska, A.-K.; Szadkowska, M.; Kozlowski, L.P.; Wyrzykowski, D.; Makowska, J.; Waters, J.J.; Swift, S.M.; Donovan, D.M.; et al. Novel Lytic Enzyme of Prophage Origin from Clostridium botulinum E3 Strain Alaska E43 with Bactericidal Activity against Clostridial Cells. Int. J. Mol. Sci. 2021, 22, 9536. https://doi.org/10.3390/ijms22179536
Morzywolek A, Plotka M, Kaczorowska A-K, Szadkowska M, Kozlowski LP, Wyrzykowski D, Makowska J, Waters JJ, Swift SM, Donovan DM, et al. Novel Lytic Enzyme of Prophage Origin from Clostridium botulinum E3 Strain Alaska E43 with Bactericidal Activity against Clostridial Cells. International Journal of Molecular Sciences. 2021; 22(17):9536. https://doi.org/10.3390/ijms22179536
Chicago/Turabian StyleMorzywolek, Agnieszka, Magdalena Plotka, Anna-Karina Kaczorowska, Monika Szadkowska, Lukasz P. Kozlowski, Dariusz Wyrzykowski, Joanna Makowska, Jerel J. Waters, Steven M. Swift, David M. Donovan, and et al. 2021. "Novel Lytic Enzyme of Prophage Origin from Clostridium botulinum E3 Strain Alaska E43 with Bactericidal Activity against Clostridial Cells" International Journal of Molecular Sciences 22, no. 17: 9536. https://doi.org/10.3390/ijms22179536
APA StyleMorzywolek, A., Plotka, M., Kaczorowska, A.-K., Szadkowska, M., Kozlowski, L. P., Wyrzykowski, D., Makowska, J., Waters, J. J., Swift, S. M., Donovan, D. M., & Kaczorowski, T. (2021). Novel Lytic Enzyme of Prophage Origin from Clostridium botulinum E3 Strain Alaska E43 with Bactericidal Activity against Clostridial Cells. International Journal of Molecular Sciences, 22(17), 9536. https://doi.org/10.3390/ijms22179536







