Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs
Abstract
:1. Introduction
2. Results
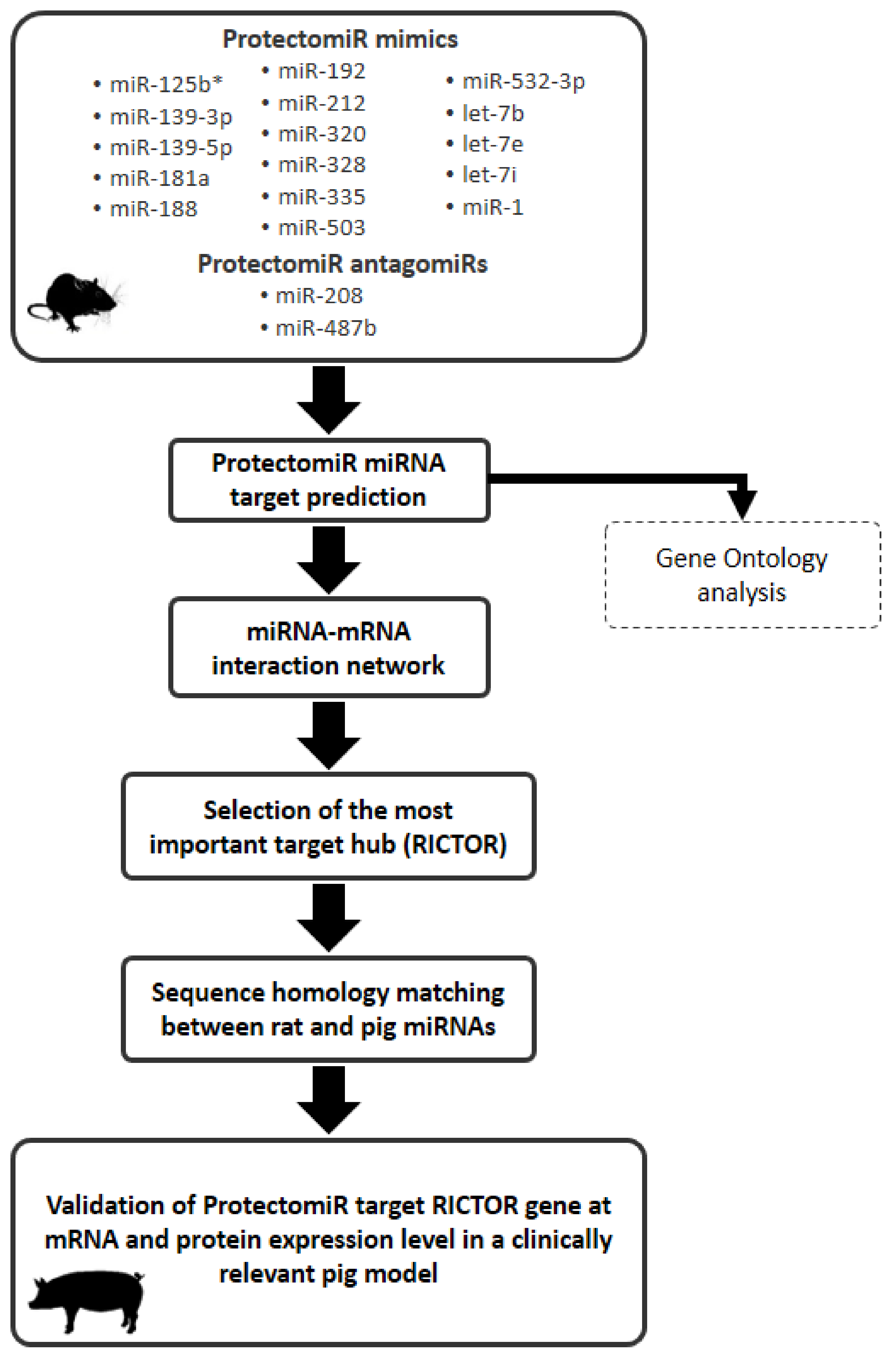
2.1. microRNA Target Prediction and ProtectomiR microRNA-mRNA Target Interaction Network
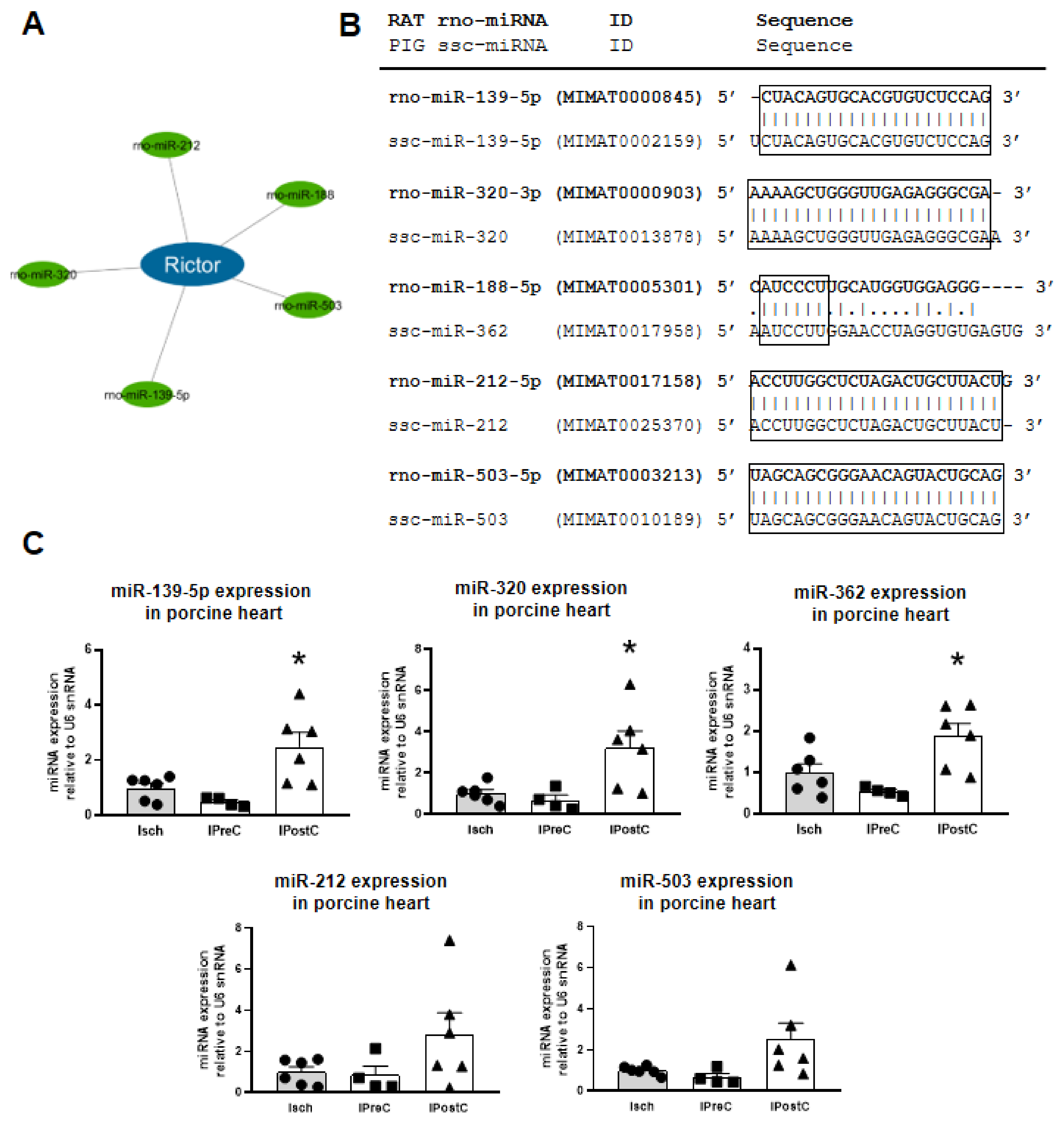
2.2. Selection of the Most Important Target Hub
2.3. Rat–Pig microRNA Homology Matching
2.4. Expression of Rictor-Targeting microRNAs in Myocardium of a Clinically Relevant Closed Chest Porcine Model
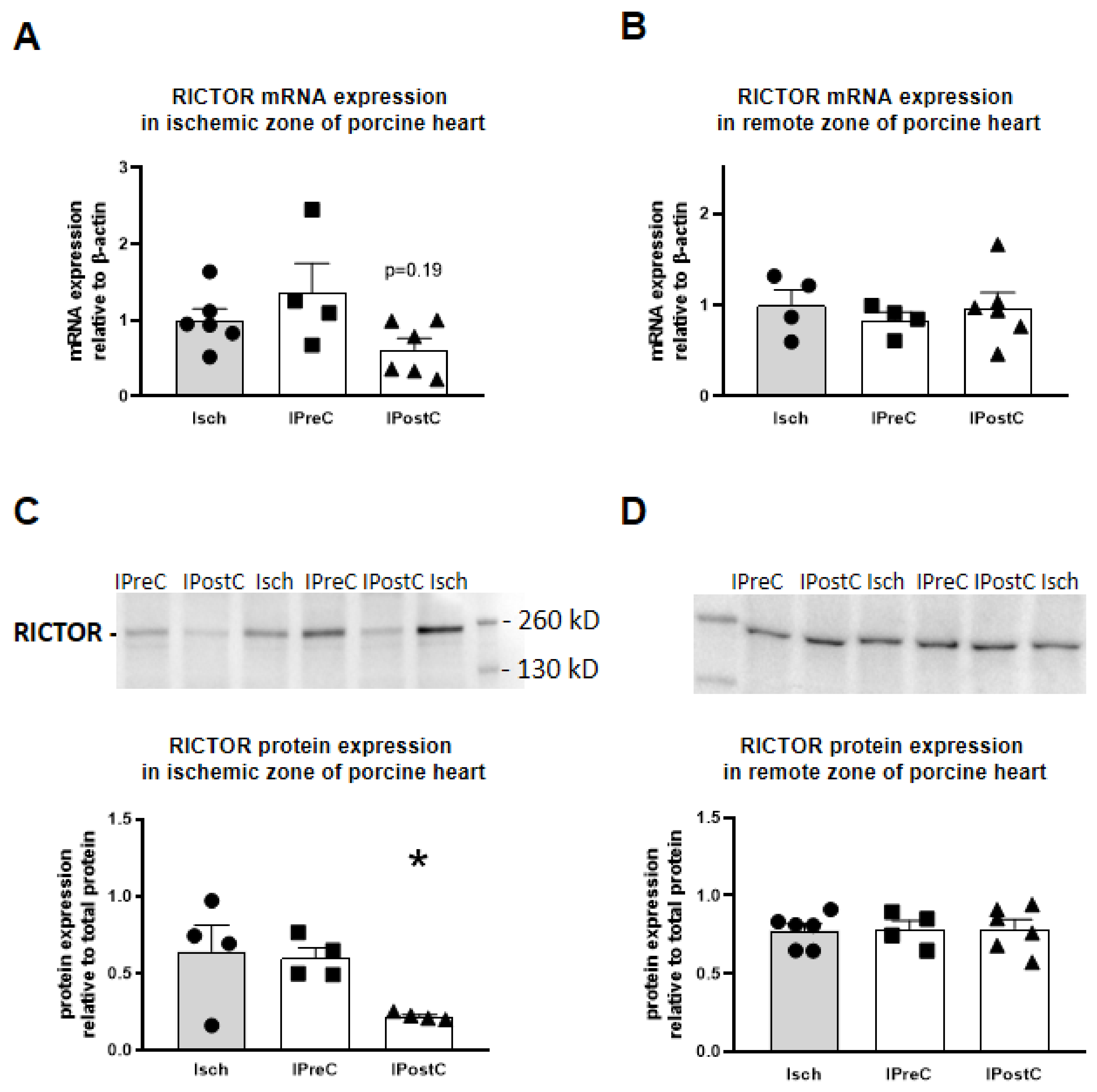
2.5. Rictor mRNA Expression
2.6. RICTOR Protein Expression
2.7. Phosphorylation Level of mTORC2 Downstream Mediators and Expression of Heat Shock Proteins in the Ischemic Zone of Myocardium
2.8. Gene Ontology Enrichment Analysis of the Interaction Network
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis of the ProtectomiR–mRNA Target Interaction Network
4.2. Cross-Species Rat–Pig microRNA Similarity Matching
4.3. Porcine Myocardial Tissue Samples
- Ischemia-reperfusion group (Isch): 90 min myocardial ischemia was induced with the balloon occlusion of the left anterior descending (LAD) coronary artery followed by 3 h of reperfusion (n = 6, except for ischemic zone western blot n = 4. Difference in group size was due to availability of samples).
- Ischemic preconditioned group (IPreC): 3 × 5 min myocardial ischemia was applied before the 90 min LAD occlusion followed by 3 h of reperfusion (n = 4, except for HSP western blots n = 3, difference in group size was due to availability of samples).
- Ischemic postconditioned group (IPostC): 6 × 30 s myocardial ischemia was applied after the 90 min LAD occlusion, at the start of the 3 h reperfusion (n = 6, except for ischemic zone western blots n = 4. difference in group size was due to availability of samples).
4.4. Total RNA Isolation
4.5. microRNA cDNA Synthesis, and qRT-PCR
4.6. mRNA cDNA Synthesis, and qRT-PCR
4.7. Western Blots
4.8. Gene Ontology Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Statement
References
- Hausenloy, D.J.; Garcia-Dorado, D.; Bøtker, H.E.; Davidson, S.M.; Downey, J.; Engel, F.B.; Jennings, R.; Lecour, S.; Leor, J.; Madonna, R.; et al. Novel targets and future strategies for acute cardioprotection: Position Paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc. Res. 2017, 113, 564–585. [Google Scholar] [CrossRef] [Green Version]
- Ferdinandy, P.; Hausenloy, D.J.; Heusch, G.; Baxter, G.F.; Schulz, R. Interaction of risk factors, comorbidities, and comedications with ischemia/reperfusion injury and cardioprotection by preconditioning, postconditioning, and remote conditioning. Pharmacol. Rev. 2014, 66, 1142–1174. [Google Scholar] [CrossRef]
- Perrino, C.; Barabási, A.L.; Condorelli, G.; Davidson, S.M.; De Windt, L.; Dimmeler, S.; Engel, F.B.; Hausenloy, D.J.; Hill, J.A.; Van Laake, L.W.; et al. Epigenomic and transcriptomic approaches in the post-genomic era: Path to novel targets for diagnosis and therapy of the ischaemic heart? Position Paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc. Res. 2017, 113, 725–736. [Google Scholar] [CrossRef]
- Ong, S.B.; Katwadi, K.; Kwek, X.Y.; Ismail, N.I.; Chinda, K.; Ong, S.G.; Hausenloy, D.J. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin. Ther. Targets 2018, 22, 247–261. [Google Scholar] [CrossRef]
- Varga, Z.V.; Zvara, A.; Faragó, N.; Kocsis, G.F.; Pipicz, M.; Gáspár, R.; Bencsik, P.; Görbe, A.; Csonka, C.; Puskás, L.G.; et al. MicroRNAs associated with ischemia-reperfusion injury and cardioprotection by ischemic pre- and postconditioning: ProtectomiRs. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H216–H227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.K.; Kafert-Kasting, S.; Thum, T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ. Res. 2020, 126, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.M.; Ferdinandy, P.; Andreadou, I.; Bøtker, H.E.; Heusch, G.; Ibáñez, B.; Ovize, M.; Schulz, R.; Yellon, D.M.; Hausenloy, D.J.; et al. Multitarget Strategies to Reduce Myocardial Ischemia/Reperfusion Injury: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2019, 73, 89–99. [Google Scholar] [CrossRef]
- Sághy, É.; Vörös, I.; Ágg, B.; Kiss, B.; Koncsos, G.; Varga, Z.V.; Görbe, A.; Giricz, Z.; Schulz, R.; Ferdinandy, P. Cardiac miRNA Expression and their mRNA Targets in a Rat Model of Prediabetes. Int. J. Mol. Sci. 2020, 21, 2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bencsik, P.; Kiss, K.; Ágg, B.; Baán, J.A.; Ágoston, G.; Varga, A.; Gömöri, K.; Mendler, L.; Faragó, N.; Zvara, Á.; et al. Sensory Neuropathy Affects Cardiac miRNA Expression Network Targeting IGF-1, SLC2a-12, EIF-4e, and ULK-2 mRNAs. Int. J. Mol. Sci. 2019, 20, 991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ágg, B.; Baranyai, T.; Makkos, A.; Vető, B.; Faragó, N.; Zvara, Á.; Giricz, Z.; Veres, D.V.; Csermely, P.; Arányi, T.; et al. MicroRNA interactome analysis predicts post-transcriptional regulation of ADRB2 and PPP3R1 in the hypercholesterolemic myocardium. Sci. Rep. 2018, 8, 10134. [Google Scholar] [CrossRef]
- Schreckenberg, R.; Klein, J.; Kutsche, H.S.; Schulz, R.; Gömöri, K.; Bencsik, P.; Benczik, B.; Ágg, B.; Sághy, É.; Ferdinandy, P.; et al. Ischaemic post-conditioning in rats: Responder and non-responder differ in transcriptome of mitochondrial proteins. J. Cell. Mol. Med. 2020, 24, 5528–5541. [Google Scholar] [CrossRef] [Green Version]
- Baehr, A.; Klymiuk, N.; Kupatt, C. Evaluating Novel Targets of Ischemia Reperfusion Injury in Pig Models. Int. J. Mol. Sci. 2019, 20, 4749. [Google Scholar] [CrossRef] [Green Version]
- Baranyai, T.; Giricz, Z.; Varga, Z.V.; Koncsos, G.; Lukovic, D.; Makkos, A.; Sarkozy, M.; Pavo, N.; Jakab, A.; Czimbalmos, C.; et al. In vivo MRI and ex vivo histological assessment of the cardioprotection induced by ischemic preconditioning, postconditioning and remote conditioning in a closed-chest porcine model of reperfused acute myocardial infarction: Importance of microvasculature. J. Transl. Med. 2017, 15, 67. [Google Scholar] [CrossRef] [Green Version]
- Ellenbroek, G.H.; van Hout, G.P.; Timmers, L.; Doevendans, P.A.; Pasterkamp, G.; Hoefer, I.E. Primary Outcome Assessment in a Pig Model of Acute Myocardial Infarction. J. Vis. Exp. JoVE 2016, 116, 54021. [Google Scholar] [CrossRef] [Green Version]
- Koudstaal, S.; Jansen of Lorkeers, S.; Gho, J.M.; van Hout, G.P.; Jansen, M.S.; Gründeman, P.F.; Pasterkamp, G.; Doevendans, P.A.; Hoefer, I.E.; Chamuleau, S.A. Myocardial infarction and functional outcome assessment in pigs. J. Vis. Exp. JoVE 2014, 86, 51269. [Google Scholar] [CrossRef] [Green Version]
- Chilukoti, R.K.; Lendeckel, J.; Darm, K.; Bukowska, A.; Goette, A.; Sühling, M.; Utpatel, K.; Peters, B.; Homuth, G.; Völker, U.; et al. Integration of “omics” techniques: Dronedarone affects cardiac remodeling in the infarction border zone. Exp. Biol. Med. 2018, 243, 895–910. [Google Scholar] [CrossRef]
- Makkos, A.; Ágg, B.; Petrovich, B.; Varga, Z.V.; Görbe, A.; Ferdinandy, P. Systematic review and network analysis of microRNAs involved in cardioprotection against myocardial ischemia/reperfusion injury and infarction: Involvement of redox signalling. Free. Radic. Biol. Med. 2021, 172, 237–251. [Google Scholar] [CrossRef]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef] [Green Version]
- Tatebe, H.; Shiozaki, K. Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules 2017, 7, 77. [Google Scholar] [CrossRef] [Green Version]
- Shende, P.; Xu, L.; Morandi, C.; Pentassuglia, L.; Heim, P.; Lebboukh, S.; Berthonneche, C.; Pedrazzini, T.; Kaufmann, B.A.; Hall, M.N.; et al. Cardiac mTOR complex 2 preserves ventricular function in pressure-overload hypertrophy. Cardiovasc. Res. 2016, 109, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Yano, T.; Ferlito, M.; Aponte, A.; Kuno, A.; Miura, T.; Murphy, E.; Steenbergen, C. Pivotal role of mTORC2 and involvement of ribosomal protein S6 in cardioprotective signaling. Circ. Res. 2014, 114, 1268–1280. [Google Scholar] [CrossRef] [Green Version]
- Lukovic, D.; Gugerell, A.; Zlabinger, K.; Winkler, J.; Pavo, N.; Baranyai, T.; Giricz, Z.; Varga, Z.V.; Riesenhuber, M.; Spannbauer, A.; et al. Transcriptional Alterations by Ischaemic Postconditioning in a Pig Infarction Model: Impact on Microvascular Protection. Int. J. Mol. Sci. 2019, 20, 344. [Google Scholar] [CrossRef] [Green Version]
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef]
- Martin, J.; Masri, J.; Bernath, A.; Nishimura, R.N.; Gera, J. Hsp70 associates with Rictor and is required for mTORC2 formation and activity. Biochem. Biophys. Res. Commun. 2008, 372, 578–583. [Google Scholar] [CrossRef] [Green Version]
- Sato, M.; Evans, B.A.; Sandström, A.L.; Chia, L.Y.; Mukaida, S.; Thai, B.S.; Nguyen, A.; Lim, L.; Tan, C.Y.R.; Baltos, J.A.; et al. α(1A)-Adrenoceptors activate mTOR signalling and glucose uptake in cardiomyocytes. Biochem. Pharmacol. 2018, 148, 27–40. [Google Scholar] [CrossRef]
- Wang, S.; Amato, K.R.; Song, W.; Youngblood, V.; Lee, K.; Boothby, M.; Brantley-Sieders, D.M.; Chen, J. Regulation of endothelial cell proliferation and vascular assembly through distinct mTORC2 signaling pathways. Mol. Cell. Biol. 2015, 35, 1299–1313. [Google Scholar] [CrossRef] [Green Version]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [Green Version]
- Hsu, S.D.; Tseng, Y.T.; Shrestha, S.; Lin, Y.L.; Khaleel, A.; Chou, C.H.; Chu, C.F.; Huang, H.Y.; Lin, C.M.; Ho, S.Y.; et al. miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014, 42, D78–D85. [Google Scholar] [CrossRef] [Green Version]
- Ágg, B.; Császár, A.; Szalay-Bekő, M.; Veres, D.V.; Mizsei, R.; Ferdinandy, P.; Csermely, P.; Kovács, I.A. The EntOptLayout Cytoscape plug-in for the efficient visualization of major protein complexes in protein-protein interaction and signalling networks. Bioinformatics 2019, 35, 4490–4492. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Attrill, H.; Gaudet, P.; Huntley, R.P.; Lovering, R.C.; Engel, S.R.; Poux, S.; Van Auken, K.M.; Georghiou, G.; Chibucos, M.C.; Berardini, T.Z.; et al. Annotation of gene product function from high-throughput studies using the Gene Ontology. Database J. Biol. Databases Curation 2019, 2019, baz007. [Google Scholar] [CrossRef]


| Degree | Symbol | Gene Name |
|---|---|---|
| 5 | Rictor | RPTOR independent companion of MTOR, complex 2 |
| 3 | Arih1 | Ariadne RBR E3 ubiquitin protein ligase 1 |
| 3 | Cd244 | CD244 molecule |
| 3 | Crisp1 | Cysteine-rich secretory protein 1 |
| 3 | Ctbs | Chitobiase |
| 3 | E2f5 | E2F transcription factor 5 |
| 3 | Ehhadh | Enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase |
| 3 | Ets1 | ETS proto-oncogene 1, transcription factor |
| 3 | Hsd17b2 | Hydroxysteroid (17-beta) dehydrogenase 2 |
| 3 | Mapk6 | Mitogen-activated protein kinase 6 |
| 3 | Mier3 | Mesoderm induction early response 1, family member 3 |
| 3 | Pde12 | Phosphodiesterase 12 |
| 3 | Slc20a1 | Solute carrier family 20 member 1 |
| 3 | Srsf7 | Serine and arginine rich splicing factor 7 |
| 3 | Yod1 | YOD1 deubiquitinase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makkos, A.; Ágg, B.; Varga, Z.V.; Giricz, Z.; Gyöngyösi, M.; Lukovic, D.; Schulz, R.; Barteková, M.; Görbe, A.; Ferdinandy, P. Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs. Int. J. Mol. Sci. 2021, 22, 9539. https://doi.org/10.3390/ijms22179539
Makkos A, Ágg B, Varga ZV, Giricz Z, Gyöngyösi M, Lukovic D, Schulz R, Barteková M, Görbe A, Ferdinandy P. Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs. International Journal of Molecular Sciences. 2021; 22(17):9539. https://doi.org/10.3390/ijms22179539
Chicago/Turabian StyleMakkos, András, Bence Ágg, Zoltán V. Varga, Zoltán Giricz, Mariann Gyöngyösi, Dominika Lukovic, Rainer Schulz, Monika Barteková, Anikó Görbe, and Péter Ferdinandy. 2021. "Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs" International Journal of Molecular Sciences 22, no. 17: 9539. https://doi.org/10.3390/ijms22179539
APA StyleMakkos, A., Ágg, B., Varga, Z. V., Giricz, Z., Gyöngyösi, M., Lukovic, D., Schulz, R., Barteková, M., Görbe, A., & Ferdinandy, P. (2021). Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs. International Journal of Molecular Sciences, 22(17), 9539. https://doi.org/10.3390/ijms22179539









