Relationships of Gut Microbiota Composition, Short-Chain Fatty Acids and Polyamines with the Pathological Response to Neoadjuvant Radiochemotherapy in Colorectal Cancer Patients
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of the Patients and Healthy Controls
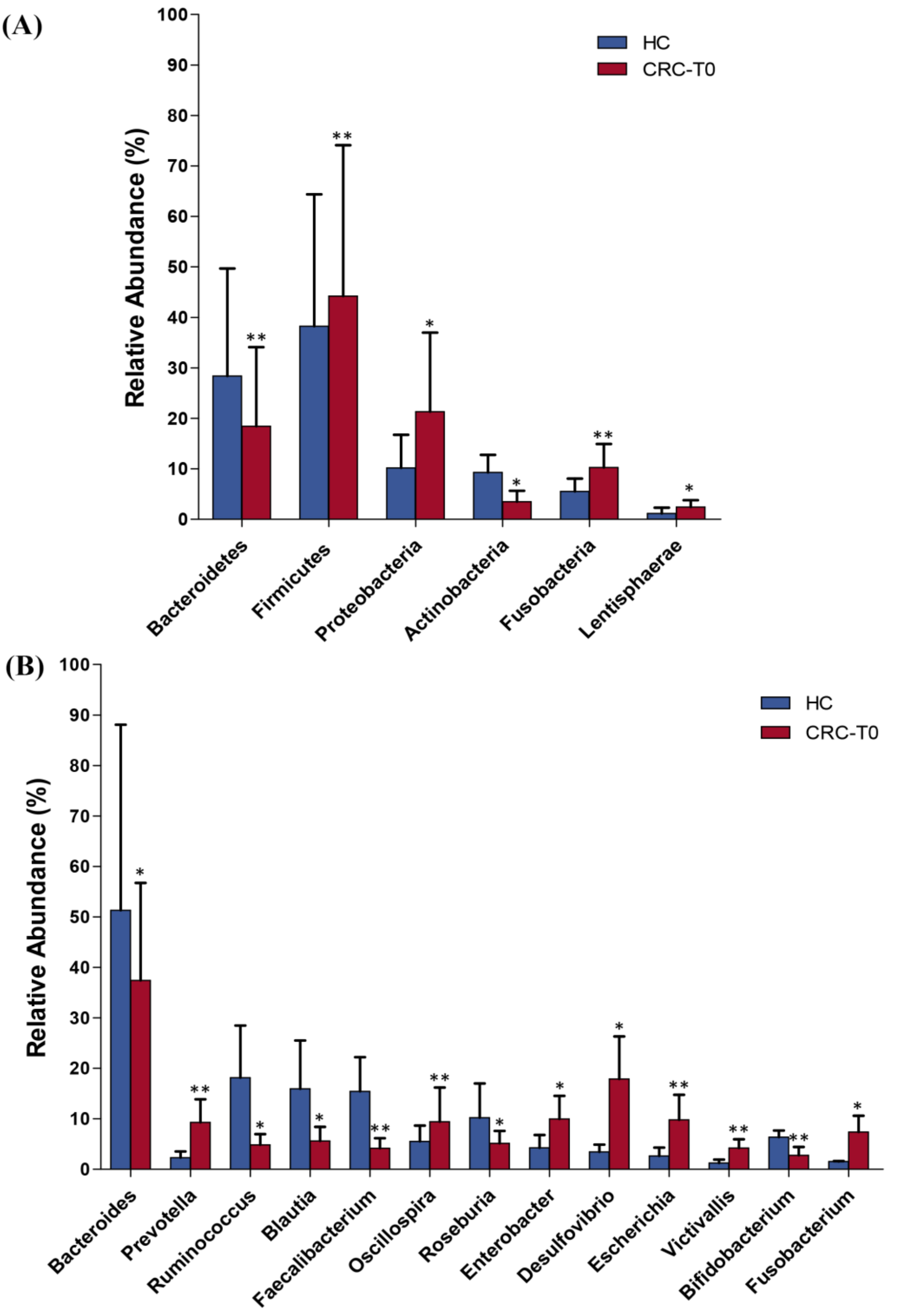
2.2. Differences in Taxonomic Composition and Diversity of Gut Microbiota between CRC Patients and Healthy Controls
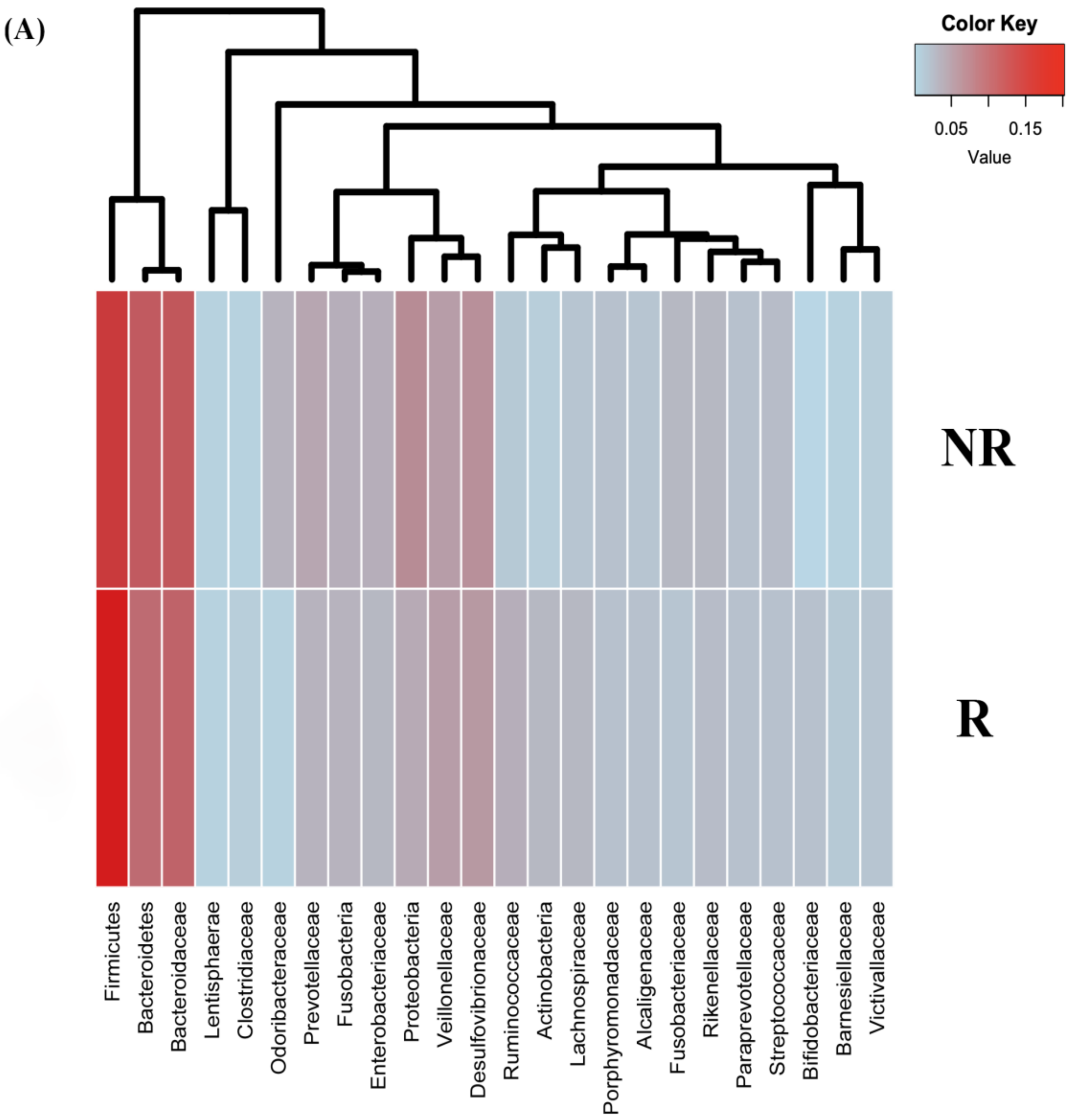
2.3. Changes in Gut Microbiota Diversity and Composition in Response to Neoadjuvant RCT Treatment in CRC Patients
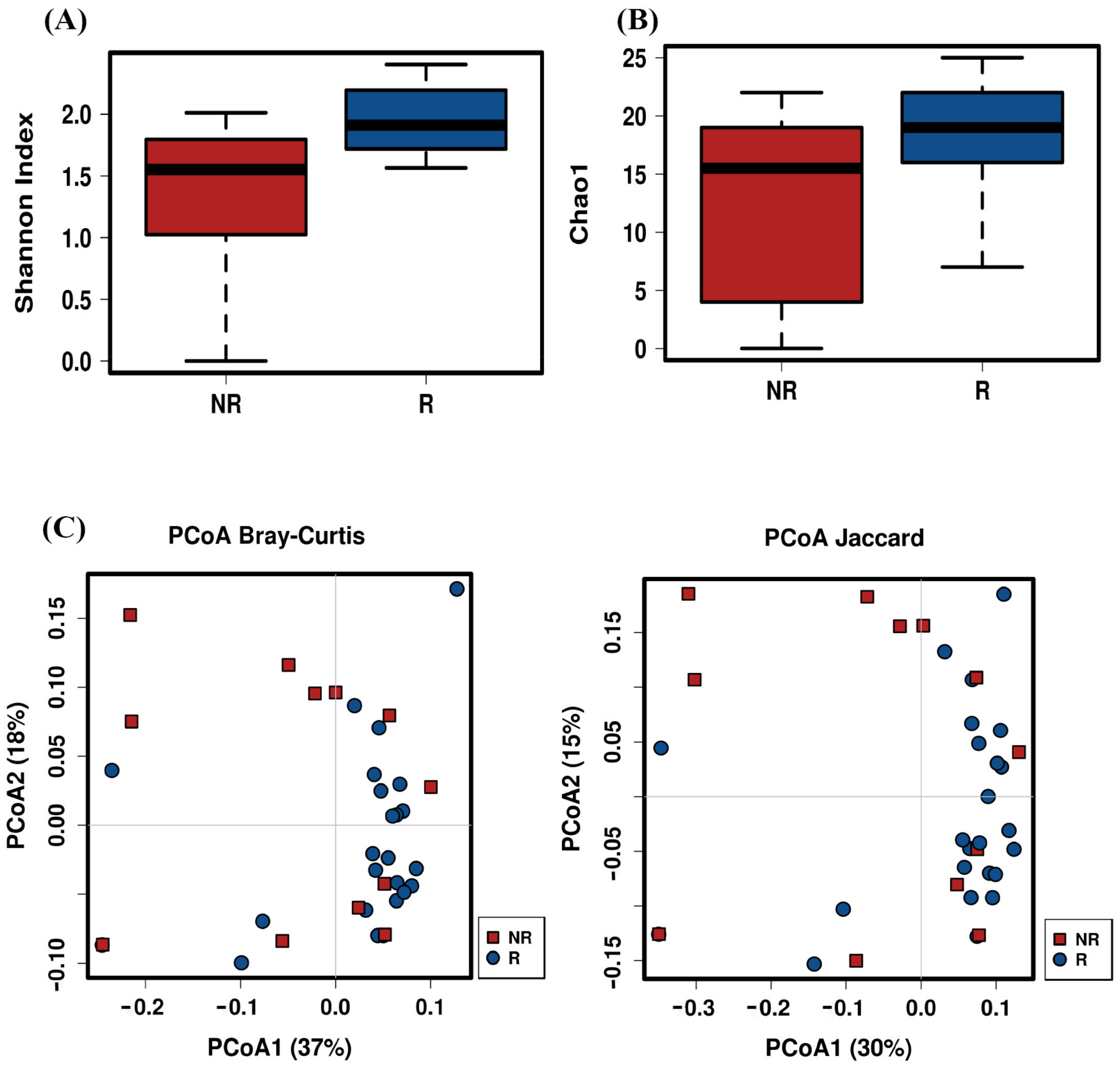
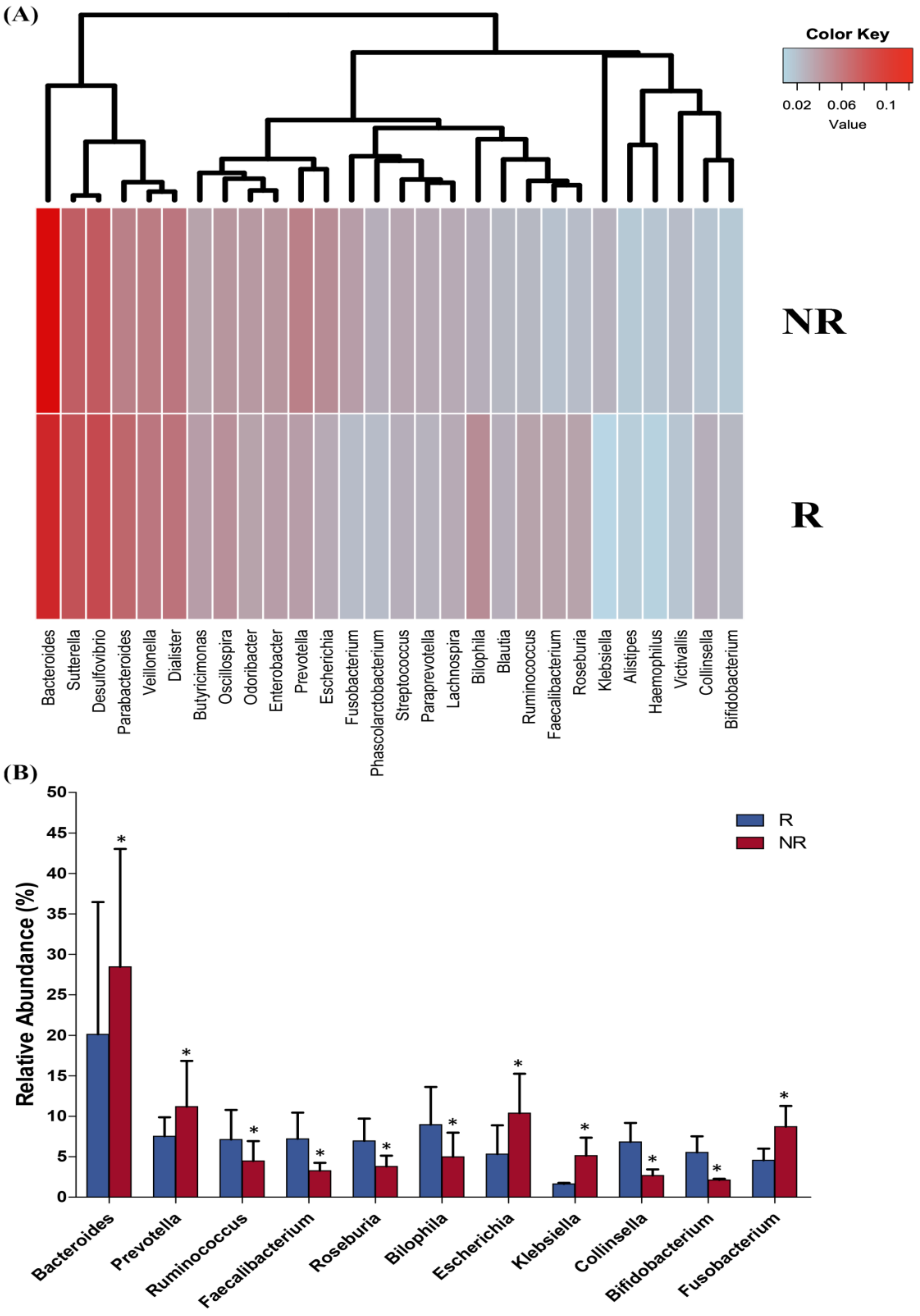
2.4. Post-Treatment Microbiota Diversity and Composition Is Associated to Clinical Response to Neoadjuvant RCT in CRC Patients
2.5. Baseline Microbiota Composition Could Predict Response to RCT Treatment in CRC Patients
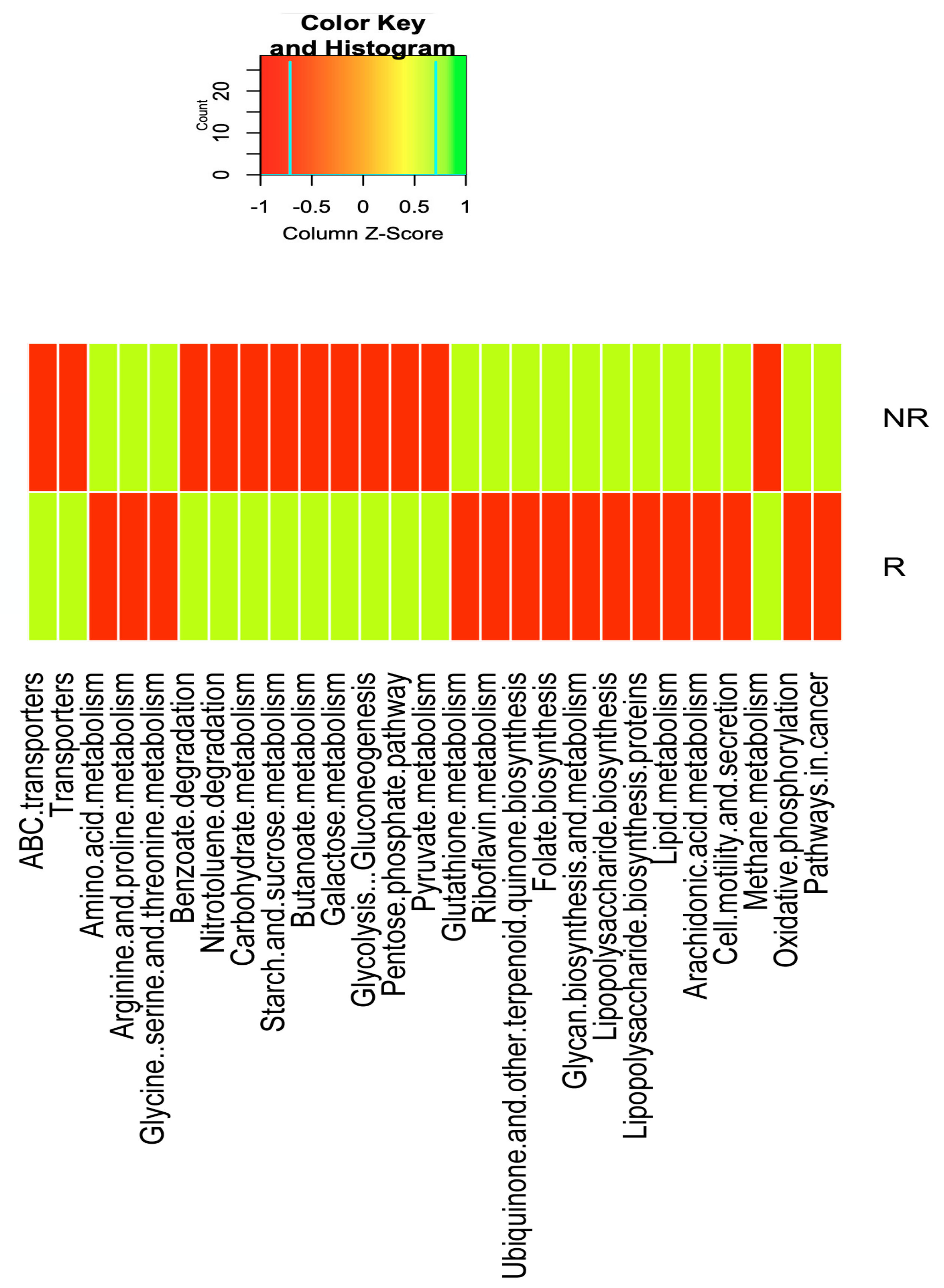
2.6. Differences in the Gut Microbiota Functions between Responder and Non-Responder
2.7. Changes in the Serum Level of Polyamines and Zonulin and Fecal Levels of SCFAs after RCT Treatment in CRC Patients
3. Discussion
4. Materials and Methods
4.1. Study Patients
4.2. Laboratory Measurements
4.3. DNA Extraction and Gut Microbiota Sequencing
4.4. Bioinformatics Analysis
4.5. Analysis of Short-Chain Fatty Acids (SCFAs) in Fecal Samples by Gas Chromatography (GC) Coupled with a Flame-Ionization Detector
4.6. Analysis of Serum Polyamine Levels by Ultra-High Performance Liquid Chromatography Tandem Mass Spectrometry (UHPLC-MS/MS)
4.7. Intestinal Permeability Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnold, D.; Lueza, B.; Douillard, J.Y.; Peeters, M.; Lenz, H.J.; Venook, A.; Heinemann, V.; Van Cutsem, E.; Pignon, J.P.; Tabernero, J.; et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann. Oncol. 2017, 28, 1713–1729. [Google Scholar] [CrossRef]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.S.M.; et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef] [Green Version]
- Yazici, C.; Wolf, P.G.; Kim, H.; Cross, T.L.; Vermillion, K.; Carroll, T.; Augustus, G.J.; Mutlu, E.; Tussing-Humphreys, L.; Brauschweig, C.; et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017, 66, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Feng, Q.; Wong, H.S.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- McQuade, J.L.; Daniel, C.R.; Helmink, B.A.; Wargo, J.A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019, 20, e77–e91. [Google Scholar] [CrossRef]
- Yi, Y.; Shen, L.; Shi, W.; Xia, F.; Zhang, H.; Wang, Y.; Zhang, J.; Wang, Y.; Sun, X.; Zhang, Z.; et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: A prospective, longitudinal study. Clin. Cancer Res. 2021, 27, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Sauer, R.; Becker, H.; Hohenberger, W.; Rödel, C.; Wittekind, C.; Fietkau, R.; Martus, P.; Tschmelitsch, J.; Hager, E.; Hess, C.F.; et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N. Engl. J. Med. 2004, 351, 1731–1740. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Gao, P.; Wang, H.; Xu, Q.; Song, Y.; Huang, X.; Sun, J.; Zhao, J.; Luo, J.; Sun, Y.; et al. What has preoperative radio(chemo)therapy brought to localized rectal cancer patients in terms of perioperative and long-term outcomes over the past decades? A systematic review and meta-analysis based on 41,121 patients. Int. J. Cancer 2017, 141, 1052–1065. [Google Scholar] [CrossRef] [Green Version]
- Pouncey, A.L.; Scott, A.J.; Alexander, J.L.; Marchesi, J.; Kinross, J. Gut microbiota, chemotherapy and the host: The influence of the gut microbiota on cancer treatment. Ecancermedicalscience 2018, 12, 868. [Google Scholar] [CrossRef] [Green Version]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. The power of small changes: Comprehensive analyses of microbial dysbiosis in breast cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 392–405. [Google Scholar] [CrossRef]
- Scott, T.A.; Quintaneiro, L.M.; Norvaisas, P.; Lui, P.P.; Wilson, M.P.; Leung, K.Y.; Herrera-Dominguez, L.; Sudiwala, S.; Pessia, A.; Clayton, P.T.; et al. Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans. Cell 2017, 169, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Sarrias, A.; Tome-Carneiro, J.; Bellesia, A.; Tomas-Barberan, F.A.; Espin, J.C. The ellagic acid-derived gut microbiota metabolite, urolithin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells. Food Funct. 2015, 6, 1460–1469. [Google Scholar] [CrossRef]
- Liu, J.; Liu, C.; Yue, J. Radiotherapy and the gut microbiome: Facts and fiction. Radiat. Oncol. 2021, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.E16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Ohara, T.; Suzutani, T. Intake of Bifidobacterium longum (BB536) and Fructo-Oligosaccharides (FOS) prevents colorectal carcinogenesis. Euroasian J. Hepatogastroenterol. 2018, 8, 11–17. [Google Scholar] [CrossRef]
- Ross, K. Gut microbial short chain fatty acids are associated with pathological complete response (pCR) after neoadjuvant chemotherapy for breast cancer. In Proceedings of the 12th European Breast Cancer Conference, Virtual Conference, 2–3 October 2020. [Google Scholar]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Taniguchi, Y.; Yoshida, A.; Nakata, K.; Nishizawa, T.; Inagawa, H.; Kohchi, C.; Soma, G. Intestinal macrophages involved in the homeostasis of the intestine have the potential for responding to LPS. Anticancer Res. 2009, 29, 4861–4865. [Google Scholar]
- Childs, A.; Mehta, D.; Gerner, E. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 2003, 60, 1394–1406. [Google Scholar] [CrossRef]
- Nakayama, Y.; Torigoe, T.; Minagawa, N.; Yamaguchi, K. The clinical usefulness of urinary N(1), N(12)-diacetylspermine (DiAcSpm) levels as a tumor marker in patients with colorectal cancer. Oncol. Lett. 2012, 3, 970–974. [Google Scholar] [CrossRef] [Green Version]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; MetaHIT Consortium; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drewes, J.L.; Housseau, F.; Sears, C.L. Sporadic colorectal cancer: Microbial contributors to disease prevention, development and therapy. Br. J. Cancer 2016, 115, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Taghinezhad-S, S.; Mohseni, A.H.; Fu, X. Intervention on gut microbiota may change the strategy for management of colorectal cancer. J. Gastroenterol. Hepatol. 2020, 36, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Heshiki, Y.; Vazquez-Uribe, R.; Li, J.; Ni, Y.; Quainoo, S.; Imamovic, L.; Li, J.; Sørensen, M.; Chow, B.K.C.; Weiss, G.J.; et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef] [PubMed]
- Saffarian, A.; Mulet, C.; Regnault, B.; Amiot, A.; Tran-Van-Nhieu, J.; Ravel, J.; Sobhani, I.; Sansonetti, P.J.; Pédron, T. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio 2019, 10, e1315–e1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, L.; Li, H.; Qin, H.; Sun, Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. OncoTargets Ther. 2017, 10, 5031–5046. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, J.; Qian, Y.; Sun, T.; Chen, H.; Chen, Z.; Sun, D.; Zhong, M.; Chen, H.; Hong, J.; et al. Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 2019, 10, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, R.Y.; Chan, C.H.; Spicer, J.D.; Rousseau, M.C.; Giannias, B.; Rousseau, S.; Ferri, L.E. LPS-induced TLR4 signaling in human colorectal cancer cells increases beta1 integrin-mediated cell adhesion and liver metastasis. Cancer Res. 2011, 71, 1989–1998. [Google Scholar] [CrossRef] [Green Version]
- Murff, H.J.; Shu, X.O.; Li, H.; Dai, Q.; Kallianpur, A.; Yang, G.; Cai, H.; Wen, W.; Gao, Y.T.; Zheng, W. A prospective study of dietary polyunsaturated fatty acids and colorectal cancer risk in Chinese women. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2283–2291. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Mann, J.R.; DuBois, R.N. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology 2005, 128, 1445–1461. [Google Scholar] [CrossRef]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a enlace with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noack, J.; Dongowski, G.; Hartmann, L.; Blaut, M. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J. Nutr. 2000, 130, 1225–1231. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.H.; Dejea, C.M.; Edler, D.; Hoang, L.T.; Santidrian, A.F.; Felding, B.H.; Ivanisevic, J.; Cho, K.; Wick, E.C.; Hechenbleikner, E.M.; et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015, 21, 891–897. [Google Scholar] [CrossRef] [Green Version]
- Mendez, R.; Kesh, K.; Arora, N.; Di Martino, L.; McAllister, F.; Merchant, N.; Banerjee, S.; Banerjee, S. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis 2020, 41, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Arruabarrena-Aristorena, A.; Zabala-Letona, A.; Carracedo, A. Oil for the cancer engine: The cross-talk between oncogenic signaling and polyamine metabolism. Sci. Adv. 2018, 4, eaar2606. [Google Scholar] [CrossRef] [Green Version]
- Bhat, M.I.; Kapila, R. Dietary metabolites derived from gut microbiota: Critical modulators of epigenetic changes in mammals. Nutr. Rev. 2017, 75, 374–389. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Amoli, H.A.; Safaei, H.G. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol. Hepatol. Bed Bench. 2018, 11, 101–109. [Google Scholar]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré., J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Martinez-Medina, M.; Abellà, C.; Busquets, D.; Sabat-Mir, M.; Duncan, S.H.; Aldeguer, X.; Flint, H.J.; Garcia-Gil, L.J. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl. Environ. Microb. 2015, 81, 7584–7592. [Google Scholar] [CrossRef] [Green Version]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de los Reyes-Gavilan, C.G. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362, fnv176. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Salcedo, R.; Worschech, A.; Cardone, M.; Jones, Y.; Gyulai, Z.; Dai, R.M.; Wang, E.; Ma, W.; Haines, D.; O’hUigin, C.; et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: Role of interleukin 18. J. Exp. Med. 2010, 207, 1625–1636. [Google Scholar] [CrossRef]
- Vaishnava, S.; Behrendt, C.L.; Ismail, A.S.; Eckmann, L.; Hooper, L.V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA 2008, 105, 20858–20863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Immerseel, F.; De Buck, J.; Boyen, F.; Bohez, L.; Pasmans, F.; Volf, J.; Sevcik, M.; Rychlik, I.; Haesebrouck, F.; Ducatelle, R. Medium-chain fatty acids decrease colonization and invasion through hilA suppression shortly after infection of chickens with Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 2004, 70, 3582–3587. [Google Scholar] [CrossRef] [Green Version]
- Sturgeon, C.; Fasano, A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers 2016, 4, e1251384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, D.P.; Rosendale, D.; Roberton, A.M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 2000, 190, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Mandard, A.M.; Dalibard, F.; Mandard, J.C.; Marnay, J.; Henry-Amar, M.; Petiot, J.F.; Roussel, A.; Jacob, J.H.; Segol, P.; Samama, G. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer 1994, 73, 2680–2686. [Google Scholar] [CrossRef]
- Ribeiro, W.R.; Vinolo, M.A.R.; Calixto, L.A.; Ferreira, C.M. Use of gas chromatography to quantify short chain fatty acids in the serum, colonic luminal content and feces of mice. Bio-Protocol 2018, 8, e3089. [Google Scholar] [CrossRef] [Green Version]
- Mendes, E.; Acetturi, B.G.; Thomas, A.M.; Martins, F.; Crisma, A.R.; Murata, G.; Braga, T.T.; Camâra, N.; Franco, A.; Setubal, J.C.; et al. Prophylactic supplementation of Bifidobacterium longum 51A protects mice from ovariectomy-induced exacerbated allergic airway inflammation and airway hyperresponsiveness. Front. Microbiol. 2017, 8, 1732. [Google Scholar] [CrossRef] [PubMed]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Liang, J. A sensitive method for the quantification of short-chain fatty acids by benzyl chloroformate derivatization combined with GC-MS. Analyst 2020, 145, 2692–2700. [Google Scholar] [CrossRef]
- Samarra, I.; Ramos-Molina, B.; Queipo-Ortuño, M.I.; Tinahones, F.J.; Arola, L.; Delpino-Rius, A.; Herrero, P.; Canela, N. Gender-related differences on polyamine metabolome in liquid biopsies by a simple and sensitive two-step liquid-liquid extraction and LC-MS/MS. Biomolecules 2019, 9, 779. [Google Scholar] [CrossRef] [Green Version]




| Healthy Controls (N = 20) | CRC-Patients (N = 40) | * p | R Patients (N = 28) | NR Patients (N = 12) | * p | |
|---|---|---|---|---|---|---|
| Age (years) | 61.42 ± 7.40 | 63.35 ± 6.97 | 0.326 | 62.93 ± 8.27 | 63.12 ± 6.34 | 0.928 |
| Gender, n (M/F) | 10/10 | 23/17 | 0.783 | 16 /12 | 7/5 | 0.780 |
| BMI (kg/m2) | 25.45 ± 3.23 | 26.42 ± 4.71 | 0.412 | 26.22 ± 4.22 | 25.92 ± 3.92 | 0.835 |
| Constipation, n (%) | 6 (20%) | 10 (25%) | 0.914 | 7 (25%) | 3 (25%) | 0.690 |
| Alcohol consumption, n (%) | 4 (13.3%) | 6 (15%) | 0.831 | 4 (14.28%) | 2 (16.16%) | 0.740 |
| Current smoking, n (%) | 9 (30%) | 15 (37.5%) | 0.774 | 11 (39.28%) | 4 (33.33%) | 0.990 |
| Biochemical data | ||||||
| Glucose (mg/dl) | 94.85 ± 19.86 | 104.79 ± 27.94 | 0.161 | 102.83 ± 26.38 | 104.15 ± 23.56 | 0.882 |
| Total cholesterol (mg/dl) | 175.2 ± 33.6 | 183.95 ± 25.71 | 0.268 | 184.17 ± 21.64 | 181.67 ± 26.12 | 0.755 |
| Triglycerides (mg/dl) | 112.67 ± 34.51 | 114.85 ± 33.62 | 0.815 | 109.25 ± 32.12 | 118.32 ± 27.12 | 0.398 |
| HDL-cholesterol (mg/dl) | 60.7 ± 15.1 | 54.83 ± 18.23 | 0.219 | 55.32 ± 16.21 | 53.89 ± 18.34 | 0.807 |
| LDL-cholesterol (mg/dl) | 107.78 ± 27.12 | 112.07 ± 33.45 | 0.621 | 109.68 ± 30.29 | 112.36 ± 33.21 | 0.805 |
| Histological variables | ||||||
| Disease stage | ||||||
| II | 22 (55%) | - | 15 (53.57%) | 7 (58.33%) | 0.945 | |
| III | 18 (45%) | - | 13 (46.42%) | 5 (41.66%) | 0.950 | |
| Tumor depth penetration (T) | ||||||
| T2–T3 | 26 (65%) | - | 18(64.28%) | 8 (66.66%) | 0.828 | |
| T4 | 14 (35%) | - | 10 (35.71%) | 4 (33.33%) | 0.832 | |
| Grade of differentiation | ||||||
| G1 | 18 (45%) | - | 12 (42.85%) | 6 (50%) | 0.944 | |
| G2 | 10 (25%) | - | 7 (25%) | 3 (25%) | 0.690 | |
| G3 | 7 (17.5%) | - | 5 (18.85%) | 2 (16.16%) | 0.806 | |
| No differentiation | 5 (12.5%) | - | 3 (10.71%) | 2 (16.66%) | 0.777 |
| R Patients (N = 28) | NR Patients (N = 12) | Between-Group Difference 1 | p2 | |
|---|---|---|---|---|
| Agmatine (ng/mL) Baseline Post-treatment Change | 0.11 ± 0.13 0.25 ± 0.24 0.14 (−0.27, −0.13) | 0.13 ± 0.15 0.17 ± 0.15 0.035 (−0.13, 0.061) | 0.025 (−0.11, 0.63) | 0.571 |
| Arginine (μg/mL) Baseline Post-treatment Change | 23.18 ± 4.20 22.82 ± 4.16 −0.36 (−1.5, 2.27) | 24.54 ± 4.76 23.10 ± 4.48 −1.43 (−1.13, 4.0) | −1.35 (−4.05, 1.35) | 0.319 |
| Ornithine (μg/mL) Baseline Post-treatment Change | 19.46 ± 5.74 20.21 ± 4.16 0.74 (−3.69, 2.19) | 23.31 ± 8.06 22.80 ± 7.55 −0.51 (−3.72, 4.74) | −3.85 (−8.07, 0,37) | 0.073 |
| N1,N12-diacetylspermine (ng/mL) Baseline Post-treatment Change | 1.08 ± 0.43 0.90 ± 0.52 −0.18 (0.017, 0.34) | 1.68 ± 1.34 1.22 ± 0.57 0.46 (−0.152, 1.07) | −0.59 (−1.20, 0.06) | 0.015 |
| N1,N8-diacetylspermidine (ng/mL) Baseline Post-treatment Change | 0.71 ± 0.26 0.74 ± 0.34 0.03 (−0.13, 0.059) | 0.99 ± 1.03 0.88 ± 0.38 −0.11 (−0.34, 0.57) | −0.28 (−0.74, 0.17) | 0.007 |
| N1-acetylspermidine (ng/mL) Baseline Post-treatment Change | 22.47 ± 7.10 23.42 ± 8.26 0.94 (−3.88, 1.99) * | 27.68 ± 13.47 28.89 ± 10.38 1.20 (−6.10, 3.68) * | −5.21 (−11.73, 1.3) | 0.021 |
| N8-acetylspermidine (ng/mL) Baseline Post-treatment Change | 14.52 ± 3.48 14.69 ± 3.39 0.16 (−0.90, 0.57) | 14.88 ± 3.27 16.10 ± 2.33 1.22 (−2.42, −0.20) * | −0.35 (−2.38, 1.67) | 0.727 |
| N1-acetylputrescine (ng/mL) Baseline Post-treatment Change | 5.04 ± 1.60 4.77 ± 1.70 −0.27 (−1.78, 1.09) | 5.92 ± 5.38 5.39 ± 3.79 −0.53 (−1.01, 3.32) | −0.88 (−3.29, 1.53) | 0.030 |
| Putrescine (ng/mL) Baseline Post-treatment Change | 8.84 ± 4.40 8.06 ± 3.89 −0.78 (−0.39, 1.96) | 7.95 ± 3.52 7.47 ± 3.09 −0.47 (−1.07, 2.02) | 0.89 (−1.49, 3.28) | 0.457 |
| Spermidine (ng/mL) Baseline Post-treatment Change | 17.14 ± 7.19 20.42 ± 12.40 3.28 (−7.42, 0.85) | 22.26 ± 12.69 20.90 ± 10.81 −1.35 (−2.01, 4.73) | −4.11 (−11.36, 1.12) | 0.106 |
| N1-acetylspermine (ng/mL) Baseline Post-treatment Change | 0.89 ± 0.33 1.19 ± 0.63 0.29 (−0.55, −0.046) | 1.48 ± 0.70 1.33 ± 0.62 −0.14 (−0.11, 0.40) | −0.58 (−0.92, −0.25) | 0.014 |
| Spermine (ng/mL) Baseline Post-treatment Change | 3.77 ± 1.30 4.80 ± 2.88 1.03 (−2.17, 0.107) * | 12.10 ± 7.85 7.35 ± 3.66 −4.74 (1.71, 7.77) * | −7.32 (−11.74, −4,89) | 0.001 |
| R Patients (N = 28) | NR Patients (N = 12) | Between-Group Difference 1 | p2 | |
|---|---|---|---|---|
| Acetic acid (mg/g) Baseline Post-treatment Change | 0.83 ± 0.39 1.04 ± 0.40 0.20 (−0.39, 0.31) * | 0.71 ± 0.15 0.77 ± 0.17 0.06 (−0.30, 0.18) | 0.26 (−0.03, 0.56) | 0.012 |
| Propionic acid (mg/g) Baseline Post-treatment Change | 1.40 ± 1.27 1.01 ± 1.10 −0.39 (−0.51, 0.59) | 2.02 ± 1.35 1.70 ± 1.52 −0.32 (−0.9, 0.36) | −0.68 (−0.86, 1.76) | 0.102 |
| Butyric acid (mg/g) Baseline Post-treatment Change | 1.37 ± 0.45 2.36 ± 1.82 0.99 (−1.2, 2.15) * | 0.93 ± 0.68 1.02 ± 1.07 0.09 (−0.65, 1.34) | 1.33 (−0.04, 2.71) | 0.016 |
| Isobutyric acid (mg/g) Baseline Post-treatment Change | 0.58 ± 0.33 0.69 ± 0.05 0.11 (0.07, 0.21) | 0.31 ± 0.33 0.44 ± 0.15 −0.13 (−0.23, 0.76) | 0.15 (0.03, 0.26) | 0.010 |
| Valeric acid (mg/g) Baseline Post-treatment Change | 0.30 ± 0.16 0.13 ± 0.07 −0.17 (−0.27, 0.39) | 0.61 ± 0.32 0.29 ± 0.19 -0.47 (−0.58, 0.76) | −0.25 (−0.38, 0.29) | 0.002 |
| Isovaleric acid (mg/g) Baseline Post-treatment Change | 0.50 ± 0.49 0.20 ± 0.13 −0.30 (−0.43, 0.31) | 0.90 ± 0.44 0.39 ± 0.24 −0.51 (0.66, 1.02) | −0.18 (−0.45, 0.29) | 0.009 |
| 4-methylvaleric acid (mg/g) Baseline Post-treatment Change | 0.13 ± 0.23 0.07 ± 0.10 −0.06 (−0.09, 0.15) | 0.37 ± 0.64 0.04 ± 0.01 −0.33 (−0.47, 0.86) | 0.20 (−0.35, 0.10) | 0.216 |
| Hexanoic acid (mg/g) Baseline Post-treatment Change | 0.15 ± 0.20 0.10 ± 0.10 −0.04 (−0.09, 0.10) | 0.11 ± 0.08 0.05 ± 0.09 −0.05 (−0.07, 0.13) | 0.05 (−0.19, 0.13) | 0.007 |
| Heptanoic acid (mg/g) Baseline Post-treatment Change | 0.09 ± 0.15 0.06 ± 0.06 −0.03 (−0.06, 0.07) | 0.07 ± 0.06 0.05 ± 0.01 −0.02 (−0.04, 0.08) | 0.02 (−0.07, 0.04) | 0.171 |
| Zonulin (ng/mL) Baseline Post-treatment Change | 257.6 ± 65.4 218.1 ± 76.4 −39.3 (−52.2, 23.9) | 272.6 ± 35.1 298.4 ± 47.5 25.2 (11.3, 37.1) | −22.2 (−37.4, 10.2) | 0.004 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Alcoholado, L.; Laborda-Illanes, A.; Otero, A.; Ordóñez, R.; González-González, A.; Plaza-Andrades, I.; Ramos-Molina, B.; Gómez-Millán, J.; Queipo-Ortuño, M.I. Relationships of Gut Microbiota Composition, Short-Chain Fatty Acids and Polyamines with the Pathological Response to Neoadjuvant Radiochemotherapy in Colorectal Cancer Patients. Int. J. Mol. Sci. 2021, 22, 9549. https://doi.org/10.3390/ijms22179549
Sánchez-Alcoholado L, Laborda-Illanes A, Otero A, Ordóñez R, González-González A, Plaza-Andrades I, Ramos-Molina B, Gómez-Millán J, Queipo-Ortuño MI. Relationships of Gut Microbiota Composition, Short-Chain Fatty Acids and Polyamines with the Pathological Response to Neoadjuvant Radiochemotherapy in Colorectal Cancer Patients. International Journal of Molecular Sciences. 2021; 22(17):9549. https://doi.org/10.3390/ijms22179549
Chicago/Turabian StyleSánchez-Alcoholado, Lidia, Aurora Laborda-Illanes, Ana Otero, Rafael Ordóñez, Alicia González-González, Isaac Plaza-Andrades, Bruno Ramos-Molina, Jaime Gómez-Millán, and María Isabel Queipo-Ortuño. 2021. "Relationships of Gut Microbiota Composition, Short-Chain Fatty Acids and Polyamines with the Pathological Response to Neoadjuvant Radiochemotherapy in Colorectal Cancer Patients" International Journal of Molecular Sciences 22, no. 17: 9549. https://doi.org/10.3390/ijms22179549
APA StyleSánchez-Alcoholado, L., Laborda-Illanes, A., Otero, A., Ordóñez, R., González-González, A., Plaza-Andrades, I., Ramos-Molina, B., Gómez-Millán, J., & Queipo-Ortuño, M. I. (2021). Relationships of Gut Microbiota Composition, Short-Chain Fatty Acids and Polyamines with the Pathological Response to Neoadjuvant Radiochemotherapy in Colorectal Cancer Patients. International Journal of Molecular Sciences, 22(17), 9549. https://doi.org/10.3390/ijms22179549







