Oestrogen Activates the MAP3K1 Cascade and β-Catenin to Promote Granulosa-like Cell Fate in a Human Testis-Derived Cell Line
Abstract
1. Introduction
2. Results
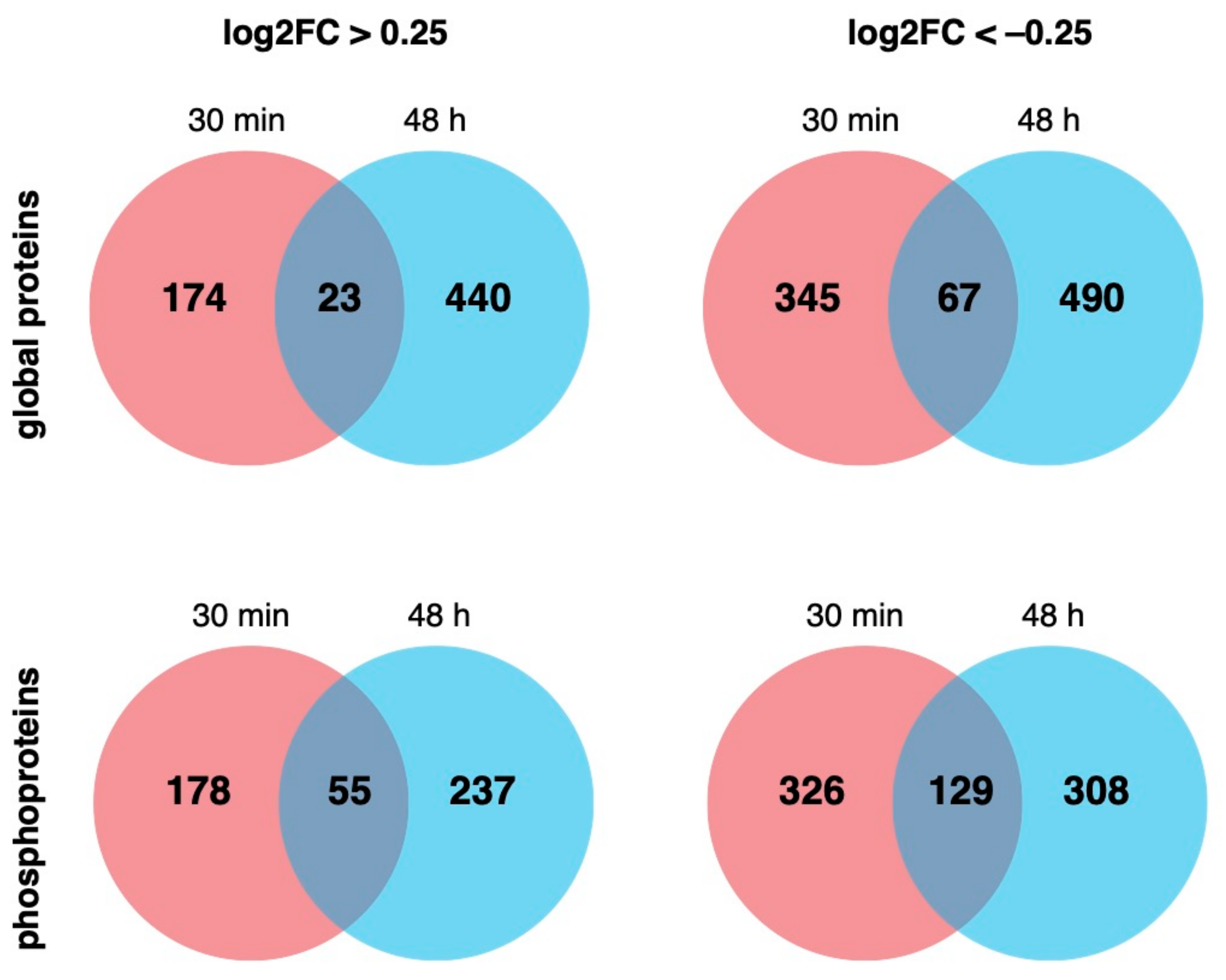
2.1. EE2 Treatment Causes Non-Genomic and Genomic Changes in NT2/D1 Cells
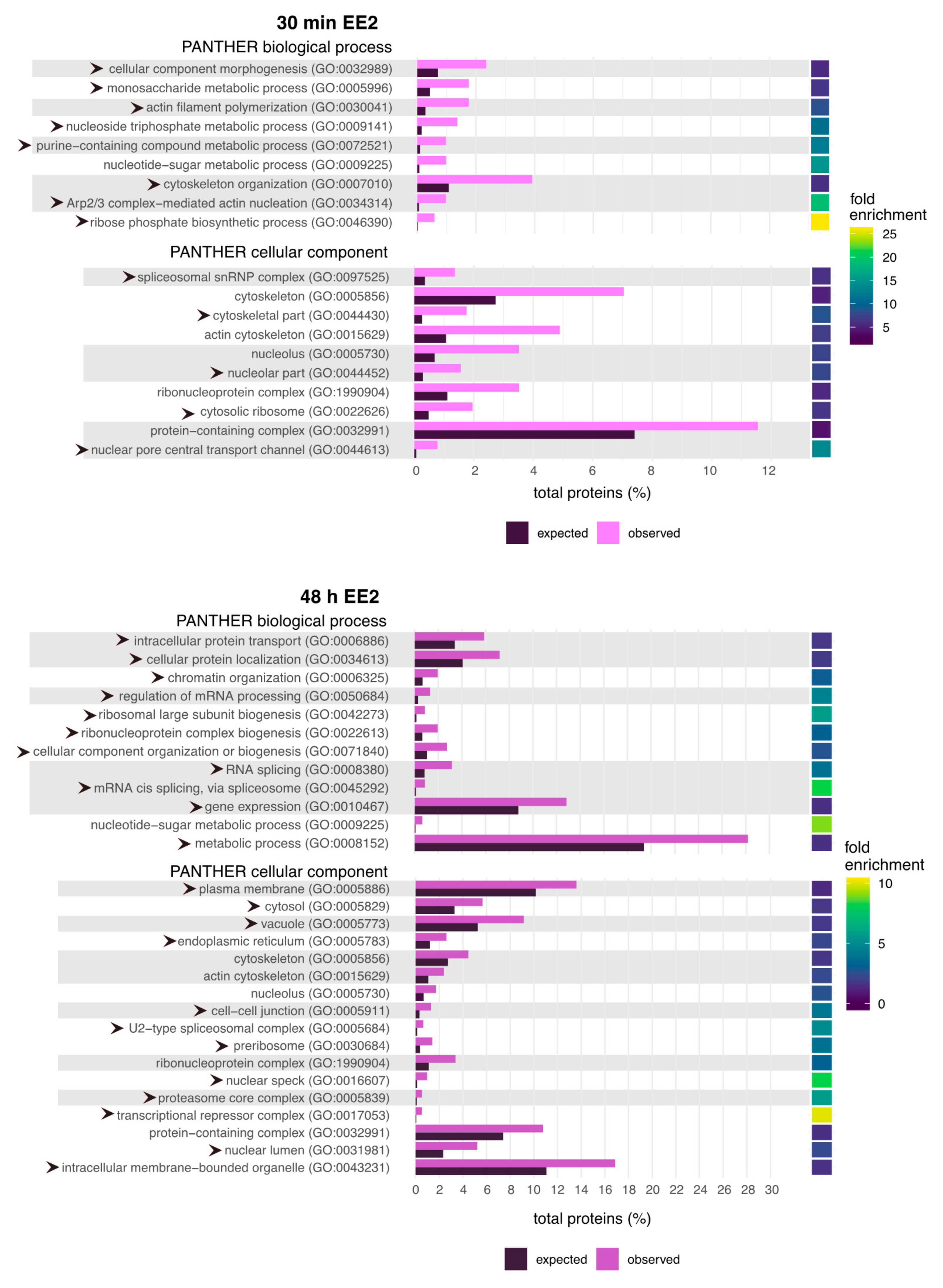
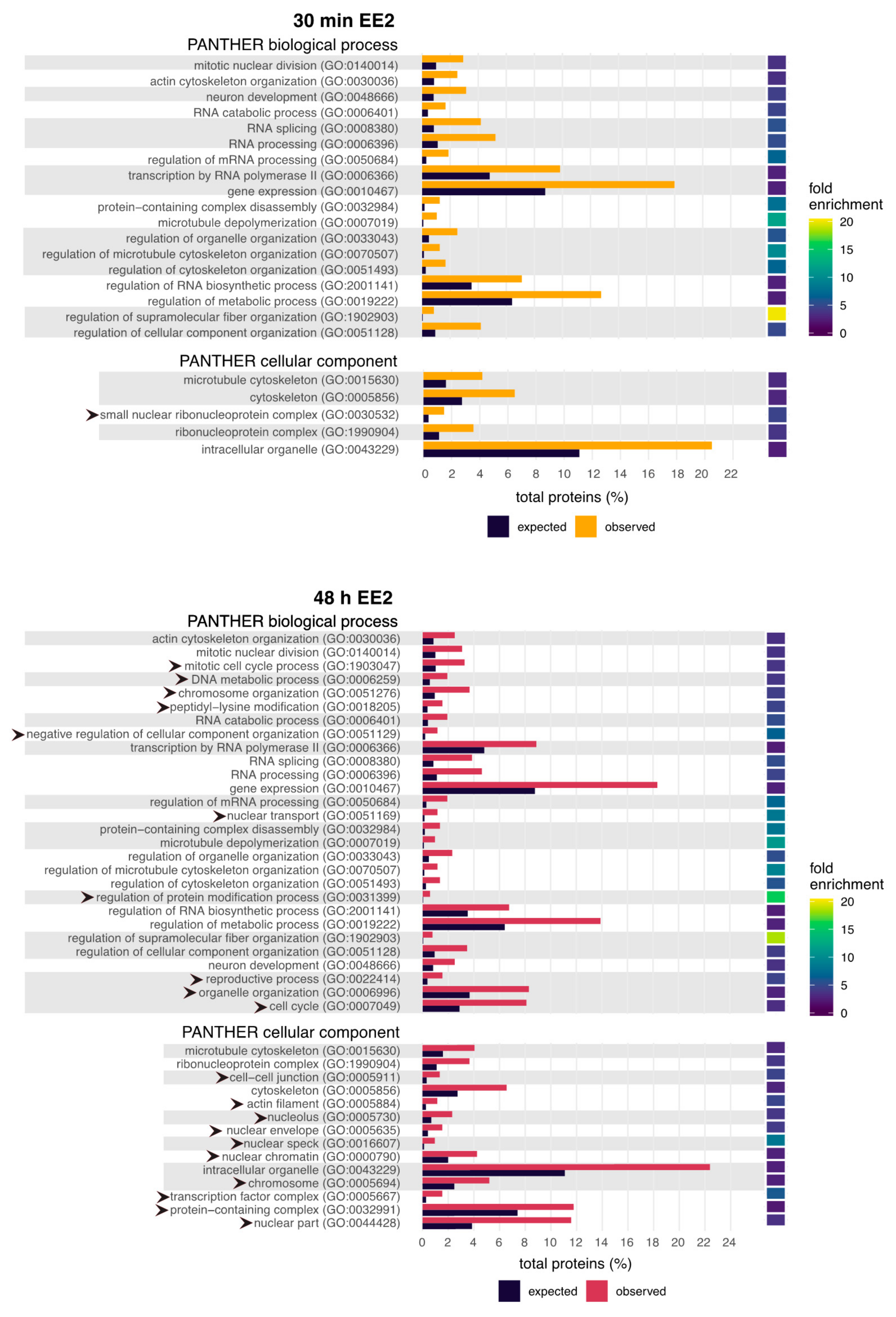
2.2. GO Terms Related to the Cytoskeleton and RNA Processing Are Enriched Following EE2 Treatment
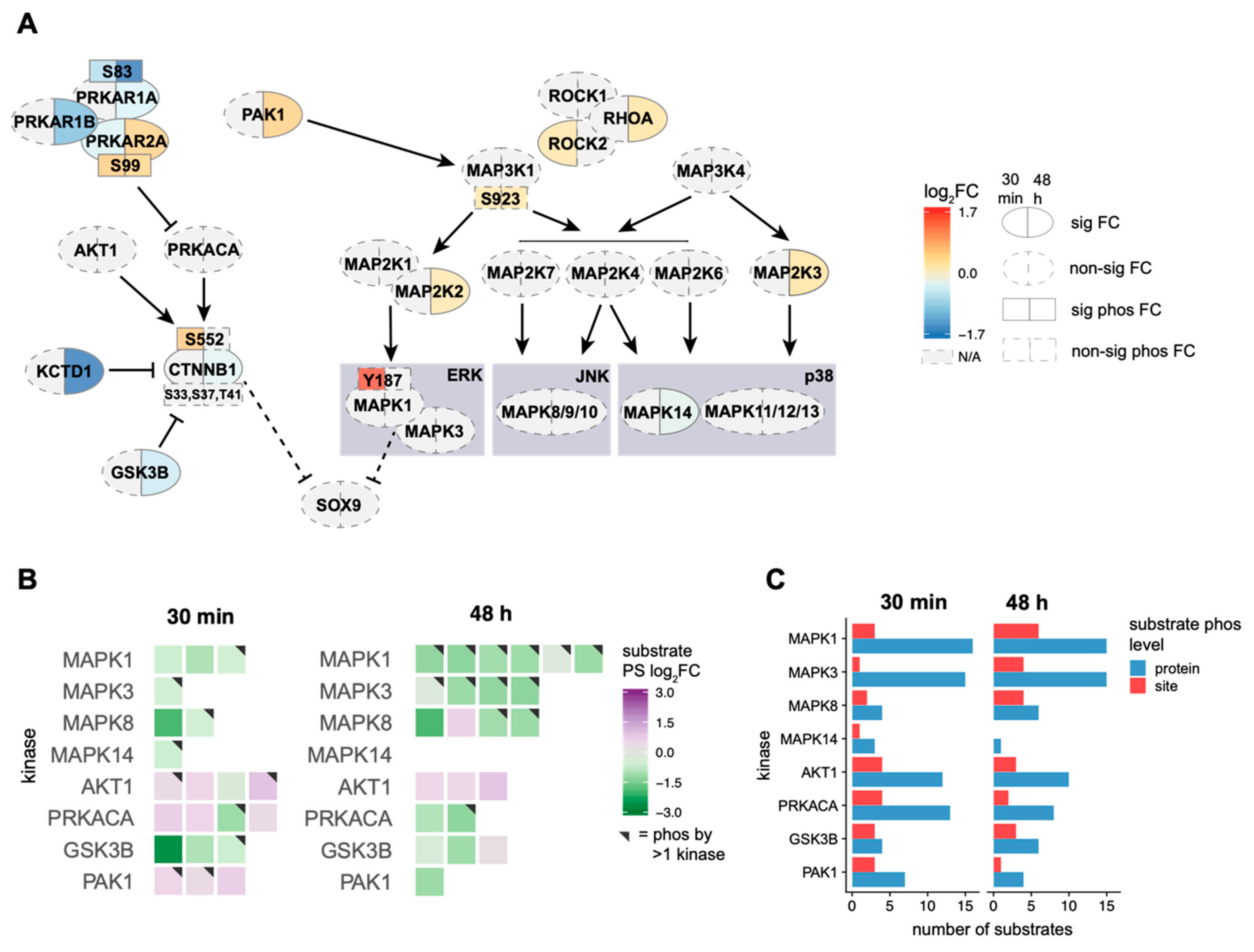
2.3. EE2 Treatment Leads to Activation of the MAP3K1 Cascade, However, Not the MAP3K4 Cascade
2.4. Oestrogen Alters the Activity of Kinases Involved in the MAP3K1/3K4 Cascades
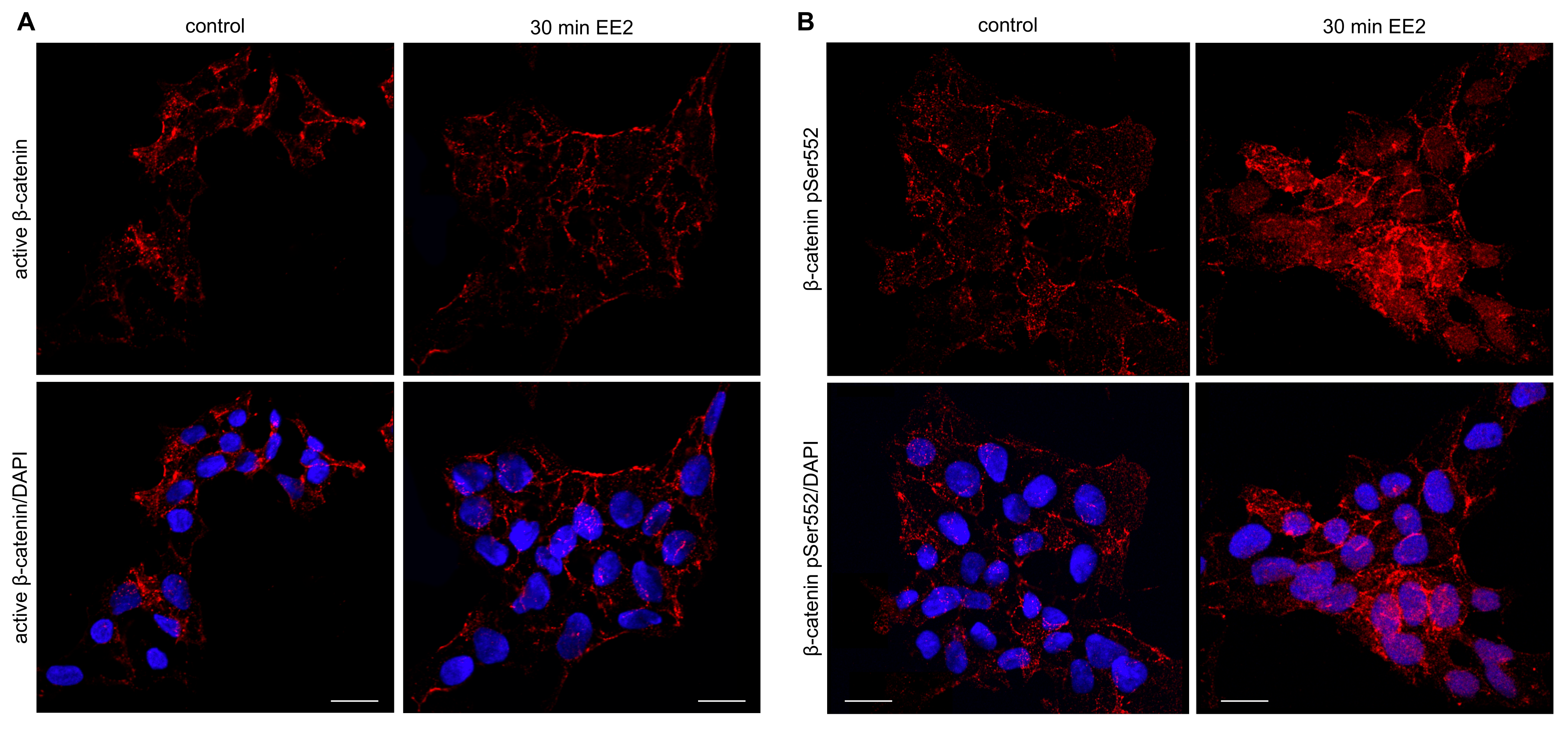
2.5. β-Catenin pSer552 Is Localized to the Nucleus Following EE2 Treatment, but Non-Phosphorylated β-Catenin Remains Unchanged
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Oestrogen Treatments, and SILAC
4.2. Protein Digestion and Phosphopeptide Enrichment
4.3. LC-MS/MS
4.4. Data Processing
4.5. Kinase-Substrate Analysis of the MAP3K1 and MAP3K4 Cascades
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ER | Oestrogen receptor |
| NT2/D1 | NTERA-2 clone D1 |
| SOX9 | Sex-determining region Y box transcription factor 9 |
| SRY | Sex-determining region Y |
| AMH | Anti-Mullerian hormone |
| FGF9 | Fibroblast growth factor |
| PTGDS | Prostaglandin D synthase |
| FOXL2 | Forkhead box L2 |
| RSPO1 | R-spondin 1 |
| WNT4 | Wnt family member 4 |
| FST | Follistatin |
| ERK1/2 | Extracellular regulated kinases 1/2 |
| MAP3K | Mitogen-activated protein kinase kinase kinase |
| RHOA | Ras homolog family member A |
| ROCK | Rho-associated coiled coil containing protein kinase |
| GSK3β | Glycogen synthase kinase 3β |
| FRAT1 | FRAT regulator of Wnt signalling pathway 1 |
| GADD45γ | Growth arrest and DNA damage-inducible protein γ |
| PKA | Protein kinase A |
| AKT | AKT serine/threonine kinase |
| PAK1 | p21 (RAC1) activated kinase 1 |
References
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.-L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Tree of Sex Consortium Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.; Crews, D. Steroid signaling and temperature-dependent sex determination—Reviewing the evidence for early action of estrogen during ovarian determination in turtles. Sem. Cell Dev. Biol. 2009, 20, 283–292. [Google Scholar] [CrossRef]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Flament, S. Sex reversal in amphibians. Sex. Dev. 2016, 10, 267–278. [Google Scholar] [CrossRef]
- Scheib, D. Effects and role of estrogens in avian gonadal differentiation. Differentiation 1983, 23, S87–S92. [Google Scholar]
- Jessl, L.; Lenz, R.; Massing, F.G.; Scheider, J.; Oehlmann, J. Effects of estrogens and antiestrogens on gonadal sex differentiation and embryonic development in the domestic fowl (Gallus gallus domesticus). PeerJ 2018, 6, e5094. [Google Scholar] [CrossRef]
- Guioli, S.; Zhao, D.; Nandi, S.; Clinton, M.; Lovell-Badge, R. Oestrogen in the chick embryo can induce chromosomally male ZZ left gonad epithelial cells to form an ovarian cortex that can support oogenesis. Development 2020, 147, dev181693. [Google Scholar] [CrossRef]
- Burns, R.K. Experimental reversal of sex in the gonads of the opossum didelphis virginiana. Proc. Natl. Acad. Sci. USA 1955, 41, 669–676. [Google Scholar] [CrossRef]
- Coveney, D.; Shaw, G.; Renfree, M.B. Estrogen-induced gonadal sex reversal in the tammar wallaby. Biol. Reprod. 2001, 65, 613–621. [Google Scholar] [CrossRef]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.-M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef]
- Sekido, R.; Bar, I.; Narváez, V.; Penny, G.; Lovell-Badge, R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev. Biol. 2004, 274, 271–279. [Google Scholar] [CrossRef]
- Barrionuevo, F.; Bagheri-Fam, S.; Klattig, J.; Kist, R.; Taketo, M.M.; Englert, C.; Scherer, G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod. 2006, 74, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Chaboissier, M.-C.; Kobayashi, A.; Vidal, V.I.P.; Lützkendorf, S.; van de Kant, H.J.G.; Wegner, M.; de Rooij, D.G.; Behringer, R.R.; Schedl, A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 2004, 131, 1891–1901. [Google Scholar] [CrossRef]
- Morais da Silva, S.; Hacker, A.; Harley, V.; Goodfellow, P.; Swain, A.; Lovell-Badge, R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat. Genet. 1996, 14, 62–68. [Google Scholar] [CrossRef]
- Wilhelm, D.; Palmer, S.; Koopman, P. Sex determination and gonadal development in mammals. Physiol. Rev. 2007, 87, 1–28. [Google Scholar] [CrossRef]
- Chassot, A.-A.; Ranc, F.; Gregoire, E.P.; Roepers-Gajadien, H.L.; Taketo, M.M.; Camerino, G.; de Rooij, D.G.; Schedl, A.; Chaboissier, M.-C. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Gen. 2008, 17, 1264–1277. [Google Scholar] [CrossRef]
- Bernard, P.; Ryan, J.; Sim, H.; Czech, D.P.; Sinclair, A.H.; Koopman, P.; Harley, V.R. Wnt signaling in ovarian development inhibits Sf1 activation of Sox9 via the Tesco enhancer. Endocrinolology 2012, 153, 901–912. [Google Scholar] [CrossRef]
- Maatouk, D.M.; DiNapoli, L.; Alvers, A.; Parker, K.L.; Taketo, M.M.; Capel, B. Stabilization of β-catenin in XY gonads causes male-to-female sex-reversal. Hum. Mol. Gen. 2008, 17, 2949–2955. [Google Scholar] [CrossRef]
- Sim, H.; Argentaro, A.; Harley, V.R. Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol. Metab. 2008, 19, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Malki, S.; Boizet-Bonhoure, B.; Poulat, F. Shuttling of SOX proteins. Int. J. Biochem. Cell Biol. 2010, 42, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Britt, K.L.; Drummond, A.E.; Dyson, M.; Wreford, N.G.; Jones, M.E.E.; Simpson, E.R.; Findlay, J.K. The ovarian phenotype of the aromatase knockout (ArKO) mouse. J. Steroid Biochem. Mol. Biol. 2001, 79, 181–185. [Google Scholar] [CrossRef]
- Couse, J.F.; Korach, K.S. Estrogen receptor null mice: What have we learned and where will they lead us? Endocr. Rev. 1999, 20, 358–417. [Google Scholar] [CrossRef]
- Pask, A.J.; Calatayud, N.E.; Shaw, G.; Wood, W.M.; Renfree, M.B. Oestrogen blocks the nuclear entry of SOX9 in the developing gonad of a marsupial mammal. BMC Biol. 2010, 8, 113. [Google Scholar] [CrossRef]
- Stewart, M.K.; Mattiske, D.M.; Pask, A.J. Estrogen suppresses SOX9 and activates markers of female development in a human testis-derived cell line. BMC Mol. Cell Biol. 2020, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.K.; Mattiske, D.M.; Pask, A.J. Oestrogen regulates SOX9 bioavailability by rapidly activating ERK1/2 and stabilising microtubules in a human testis-derived cell line. Exp. Cell Res. 2021, 398, 112405. [Google Scholar] [CrossRef] [PubMed]
- Jessop, H.L.; Sjöberg, M.; Cheng, M.Z.; Zaman, G.; Wheeler-Jones, C.P.; Lanyon, L.E. Mechanical strain and estrogen activate estrogen receptor alpha in bone cells. J. Bone Miner. Res. 2001, 16, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Endoh, H.; Sasaki, H.; Maruyama, K.; Takeyama, K.; Waga, I.; Shimizu, T.; Kato, S.; Kawashima, H. Rapid activation of MAP kinase by estrogen in the bone cell line. Biochem. Biophys. Res. 1997, 235, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yuhanna, I.S.; Galcheva-Gargova, Z.; Karas, R.H.; Mendelsohn, M.E.; Shaul, P.W. Estrogen receptor α mediates the nongenomic activation of endothelial nitric oxide synthase by estrogen. J. Clin. Investig. 1999, 103, 401–406. [Google Scholar] [CrossRef]
- Watters, J.J.; Campbell, J.S.; Cunningham, M.J.; Krebs, E.G.; Dorsa, D.M. Rapid membrane effects of steroids in neuroblastoma cells: Effects of estrogen on mitogen activated protein kinase signaling cascade and c-fos immediate early gene transcription. Endocrinology 1997, 138, 4030–4033. [Google Scholar] [CrossRef]
- Pearlman, A.; Loke, J.; Le Caignec, C.; White, S.; Chin, L.; Friedman, A.; Warr, N.; Willan, J.; Brauer, D.; Farmer, C.; et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am. J. Hum. Genet. 2010, 87, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, A.; Huether, R.; Machado, A.Z.; Groden, M.; Liu, H.-M.; Upadhyay, K.; Vivian, O.; Gomes, N.L.; Lerario, A.M.; Nishi, M.Y.; et al. Mutations in MAP3K1 that cause 46,XY disorders of sex development disrupt distinct structural domains in the protein. Hum. Mol. Gen. 2019, 28, 1620–1628. [Google Scholar] [CrossRef]
- Loke, J.; Pearlman, A.; Radi, O.; Zuffardi, O.; Giussani, U.; Pallotta, R.; Camerino, G.; Ostrer, H. Mutations in MAP3K1 tilt the balance from SOX9/FGF9 to WNT/beta-catenin signaling. Hum. Mol. Gen. 2014, 23, 1073–1083. [Google Scholar] [CrossRef]
- Warr, N.; Carre, G.-A.; Siggers, P.; Faleato, J.V.; Brixey, R.; Pope, M.; Bogani, D.; Childers, M.; Wells, S.; Scudamore, C.L.; et al. Gadd45γ and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell 2012, 23, 1020–1031. [Google Scholar] [CrossRef]
- Mulligan, W.A.; Wegner, K.A.; Keil, K.P.; Mehta, V.; Taketo, M.M.; Vezina, C.M. Beta-catenin and estrogen signaling collaborate to drive cyclin D1 expression in developing mouse prostate. Differentiation 2017, 93, 66–71. [Google Scholar] [CrossRef]
- Ray, S.; Xu, F.; Wang, H.; Das, S.K. Cooperative control via lymphoid enhancer factor 1/T cell factor 3 and estrogen receptor-α for uterine gene regulation by estrogen. Mol. Endocrinol. 2008, 22, 1125–1140. [Google Scholar] [CrossRef]
- Xiong, W.; Zhang, L.; Liu, H.; Li, N.; Du, Y.; He, H.; Zhang, Z.; Liu, Y. E 2-mediated EMT by activation of β-catenin/Snail signalling during the development of ovarian endometriosis. J. Cell Mol. Med. 2019, 23, 8035–8045. [Google Scholar] [CrossRef] [PubMed]
- Rubinfeld, B.; Albert, I.; Porfiri, E.; Fiol, C.; Munemitsu, S.; Polakis, P. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science 1996, 272, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Hawke, D.; Zheng, Y.; Xia, Y.; Meisenhelder, J.; Nika, H.; Mills, G.B.; Kobayashi, R.; Hunter, T.; Lu, Z. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 2007, 282, 11221–11229. [Google Scholar] [CrossRef]
- Taurin, S.; Sandbo, N.; Qin, Y.; Browning, D.; Dulin, N.O. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 2006, 281, 9971–9976. [Google Scholar] [CrossRef]
- Kawagoe, J.; Ohmichi, M.; Takahashi, T.; Ohshima, C.; Mabuchi, S.; Takahashi, K.; Igarashi, H.; Mori-Abe, A.; Saitoh, M.; Du, B.; et al. Raloxifene inhibits estrogen-induced up-regulation of telomerase activity in a human breast cancer cell line. J. Biol. Chem. 2003, 278, 43363–43372. [Google Scholar] [CrossRef]
- Prossnitz, E.R. GPER modulators: Opportunity Nox on the heels of a class Akt. J. Steroid Biochem. Mol. Biol. 2018, 176, 73–81. [Google Scholar] [CrossRef]
- Bouskine, A.; Nebout, M.; Mograbi, B.; Brücker-Davis, F.; Roger, C.; Fenichel, P. Estrogens promote human testicular germ cell cancer through a membrane-mediated activation of extracellular regulated kinase and protein kinase A. Endocrinology 2008, 149, 565–573. [Google Scholar] [CrossRef]
- Tang, S.; Han, H.; Bajic, V.B. ERGDB: Estrogen responsive genes database. Nucleic Acids Res. 2004, 32, D533–D536. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Li, X.; Chen, C.; Wang, F.; Huang, W.; Liang, Z.; Xiao, Y.; Wei, K.; Wan, Z.; Hu, X.; Xiang, S.; et al. KCTD1 suppresses canonical wnt signaling pathway by enhancing β-catenin degradation. PLoS ONE 2014, 9, e94343. [Google Scholar] [CrossRef]
- Needham, E.J.; Parker, B.L.; Burykin, T.; James, D.E.; Humphrey, S.J. Illuminating the dark phosphoproteome. Sci. Signal. 2019, 12, eaau8645. [Google Scholar] [CrossRef]
- Bronson, M.W.; Hillenmeyer, S.; Park, R.W.; Brodsky, A.S. Estrogen coordinates translation and transcription, revealing a role for NRSF in human breast cancer cells. Mol. Endocrinol. 2010, 24, 1120–1135. [Google Scholar] [CrossRef][Green Version]
- Cuesta, R.; Gritsenko, M.A.; Petyuk, V.A.; Shukla, A.K.; Tsai, C.-F.; Liu, T.; McDermott, J.E.; Holz, M.K. Phosphoproteome analysis reveals estrogen-ER pathway as a modulator of mTOR activity via DEPTOR. Mol. Cell Proteom. 2019, 18, 1607–1618. [Google Scholar] [CrossRef]
- Correia, B.; Sousa, M.I.; Ramalho-Santos, J. The mTOR pathway in reproduction: From gonadal function to developmental coordination. Reproduction 2020, 159, R173–R188. [Google Scholar] [CrossRef] [PubMed]
- Paranjpe, M.; Yu, H.; Frankenberg, S.; Pask, A.J.; Shaw, G.; Renfree, M.B. Transcriptomic analysis of MAP3K1 and MAP3K4 in the developing marsupial gonad. Sex. Dev. 2019, 13, 195–204. [Google Scholar] [CrossRef]
- Woods, A.; Wang, G.; Beier, F. RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem. 2005, 280, 11626–11634. [Google Scholar] [CrossRef]
- Kumar, D.; Lassar, A.B. The transcriptional activity of Sox9 in chondrocytes is regulated by RhoA signaling and actin polymerization. Mol. Cell Biol. 2009, 29, 4262–4273. [Google Scholar] [CrossRef]
- Varea, O.; Garrido, J.J.; Dopazo, A.; Mendez, P.; Garcia-Segura, L.M.; Wandosell, F. Estradiol activates beta-catenin dependent transcription in neurons. PLoS ONE 2009, 4, e5153. [Google Scholar] [CrossRef]
- Britt, K.L.; Findlay, J.K. Regulation of the phenotype of ovarian somatic cells by estrogen. Mol. Cell Endocrinol. 2003, 202, 11–17. [Google Scholar] [CrossRef]
- Cardona-Gomez, P.; Pérez, M.; Avila, J.; Garcia-Segura, L.M.; Wandosell, F. Estradiol inhibits GSK3 and regulates interaction of estrogen receptors, GSK3, and beta-catenin in the hippocampus. Mol. Cell. Neurosci. 2004, 25, 363–373. [Google Scholar] [CrossRef]
- Knower, K.C.; Sim, H.; McClive, P.J.; Bowles, J.; Koopman, P.; Sinclair, A.H.; Harley, V.R. Characterisation of urogenital ridge gene expression in the human embryonal carcinoma cell line NT2/D1. Sex. Dev. 2007, 1, 114–126. [Google Scholar] [CrossRef]
- Knower, K.C.; Kelly, S.; Ludbrook, L.M.; Bagheri-Fam, S.; Sim, H.; Bernard, P.; Sekido, R.; Lovell-Badge, R.; Harley, V.R. Failure of SOX9 regulation in 46XY disorders of sex development with SRY, SOX9 and SF1 mutations. PLoS ONE 2011, 6, e17751. [Google Scholar] [CrossRef]
- Ong, S.-E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Adeel, M.; Song, X.; Wang, Y.; Francis, D.; Yang, Y. Environmental impact of estrogens on human, animal and plant life: A critical review. Environ. Int. 2017, 99, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Aris, A.Z.; Shamsuddin, A.S.; Praveena, S.M. Occurrence of 17α-ethynylestradiol (EE2) in the environment and effect on exposed biota: A review. Environ. Int. 2014, 69, 104–119. [Google Scholar] [CrossRef]
- Larsen, M.R.; Jensen, S.S.; Jakobsen, L.A.; Heegaard, N.H.H. Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell Proteom. 2007, 6, 1778–1787. [Google Scholar] [CrossRef]
- Wang, T.; Ma, G.; Ang, C.-S.; Korhonen, P.K.; Stroehlein, A.J.; Young, N.D.; Hofmann, A.; Chang, B.C.H.; Williamson, N.A.; Gasser, R.B. The developmental phosphoproteome of Haemonchus contortus. J. Proteom. 2020, 213, 103615. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Leeming, M.G.; O’Callaghan, S.; Licata, L.; Iannuccelli, M.; Lo Surdo, P.; Micarelli, E.; Ang, C.-S.; Nie, S.; Varshney, S.; Ameen, S.; et al. Phosphomatics: Interactive interrogation of substrate–kinase networks in global phosphoproteomics datasets. Bioinformatics 2020, 10, 1–19. [Google Scholar]
- Perfetto, L.; Briganti, L.; Calderone, A.; Cerquone Perpetuini, A.; Iannuccelli, M.; Langone, F.; Licata, L.; Marinkovic, M.; Mattioni, A.; Pavlidou, T.; et al. SIGNOR: A database of causal relationships between biological entities. Nucleic Acids Res. 2015, 44, D548–D554. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stewart, M.K.; Bernard, P.; Ang, C.-S.; Mattiske, D.M.; Pask, A.J. Oestrogen Activates the MAP3K1 Cascade and β-Catenin to Promote Granulosa-like Cell Fate in a Human Testis-Derived Cell Line. Int. J. Mol. Sci. 2021, 22, 10046. https://doi.org/10.3390/ijms221810046
Stewart MK, Bernard P, Ang C-S, Mattiske DM, Pask AJ. Oestrogen Activates the MAP3K1 Cascade and β-Catenin to Promote Granulosa-like Cell Fate in a Human Testis-Derived Cell Line. International Journal of Molecular Sciences. 2021; 22(18):10046. https://doi.org/10.3390/ijms221810046
Chicago/Turabian StyleStewart, Melanie K., Pascal Bernard, Ching-Seng Ang, Deidre M. Mattiske, and Andrew J. Pask. 2021. "Oestrogen Activates the MAP3K1 Cascade and β-Catenin to Promote Granulosa-like Cell Fate in a Human Testis-Derived Cell Line" International Journal of Molecular Sciences 22, no. 18: 10046. https://doi.org/10.3390/ijms221810046
APA StyleStewart, M. K., Bernard, P., Ang, C.-S., Mattiske, D. M., & Pask, A. J. (2021). Oestrogen Activates the MAP3K1 Cascade and β-Catenin to Promote Granulosa-like Cell Fate in a Human Testis-Derived Cell Line. International Journal of Molecular Sciences, 22(18), 10046. https://doi.org/10.3390/ijms221810046






