Selective Targeting of Human and Animal Pathogens of the Helicobacter Genus by Flavodoxin Inhibitors: Efficacy, Synergy, Resistance and Mechanistic Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
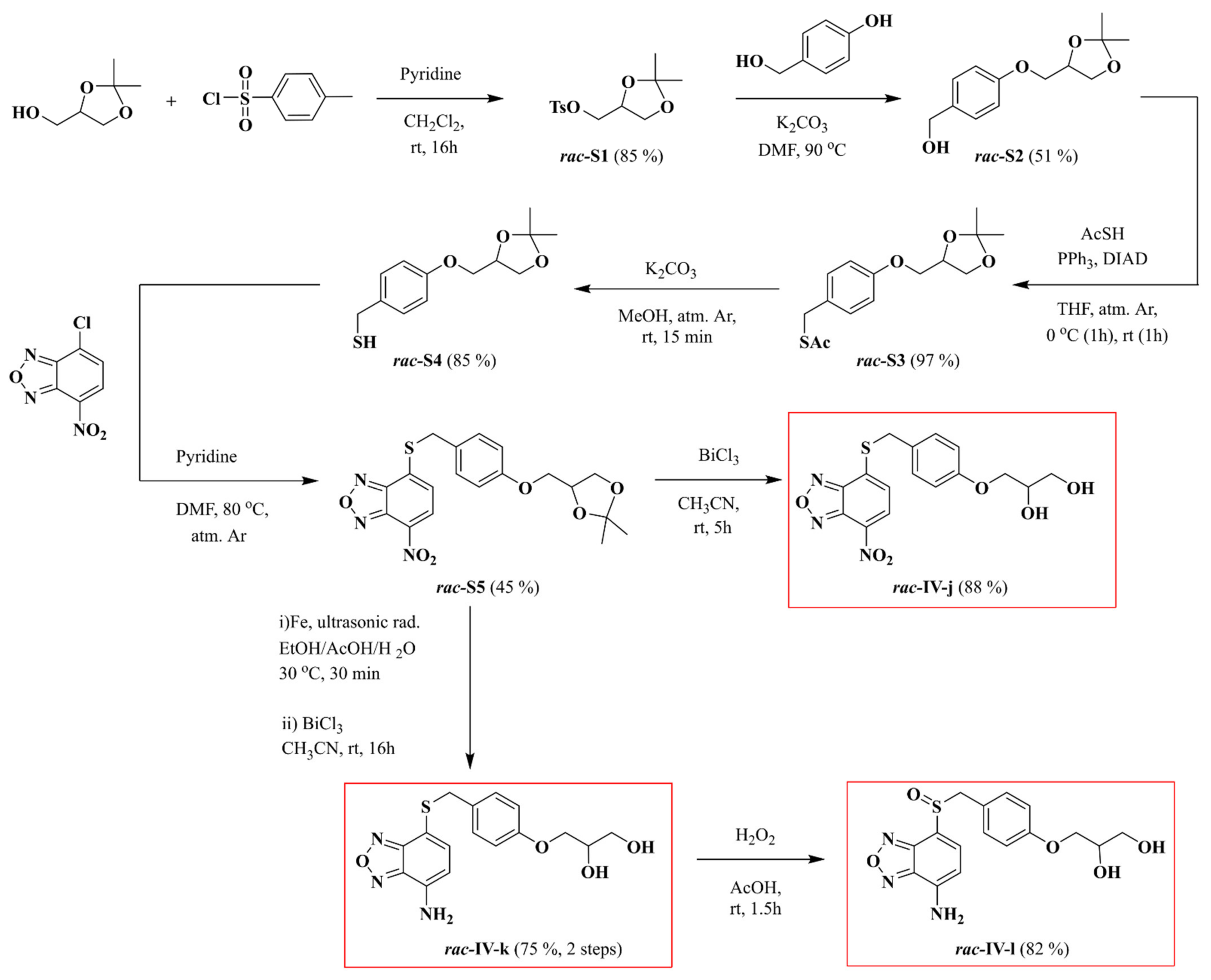
2.2. Synthesis of Compounds Rac-IV-j, Rac-IV-k and Rac-IV-l
2.2.1. rac-(2,2-Dimethyl-1,3-dioxolan-4-yl)methyl p-Toluenenesulfonate (rac-S1)
2.2.2. rac-(4-((2,2-Dimethyl-1,3-dioxolan-4-yl)methoxy)phenyl)methanol (rac-S2)
2.2.3. rac-S-(4-((2,2-Dimethyl-1,3-dioxolan-4-yl)methoxy)benzyl) Thioacetate (rac-S3)
2.2.4. rac-(4-((2,2-Dimethyl-1,3-dioxolan-4-yl)methoxy)phenyl)methanethiol (rac-S4)
2.2.5. rac-4-((4-((2,2-Dimethyl-1,3-dioxolan-4-yl)methoxy)benzyl)thio)-7-nitrobenzo[c][1,2,5]oxadiazole (rac-S5)
2.2.6. rac-3-(4-(((7-Nitrobenzo[c][1,2,5]oxadiazol-4-yl)thio)methyl)phenoxy)propane-1,2-diol (rac-IV-j)
2.2.7. rac-3-(4-(((7-Aminobenzo[c][1,2,5]oxadiazol-4-yl)thio)methyl)phenoxy)propane-1,2-diol (rac-IV-k)
2.2.8. rac-3-(4-(((7-Aminobenzo[c][1,2,5]oxadiazol-4-yl)sulfinyl)methyl)phenoxy)propane-1,2-diol (rac-IV-l)
2.3. Bacterial Strains, Culture Media and Growth Conditions
2.4. Evaluation of the Antibacterial Activity against a Diverse Microbial Panel
2.5. Cloning, Expression, Purification and Quantification of Recombinant Flavodoxin
2.6. Determination of the Hp-Fld-Binding Affinity of Inhibitors
2.7. Molecular Docking of Compounds IV, IV-a, IV-b, IV-c and IV-d to Hp-Fld
2.8. Combined Antimicrobial Effect
2.9. Generation of Spontaneous Resistant Mutants
2.10. Chequerboard Synergy Testing
3. Results
3.1. Selectivity of the Flavodoxin Inhibitors for Bacteria of the Helicobacter Genus
3.2. Probable Interaction of the Inhibitors at a Pocket near the FMN Binding Site
3.3. Prospects of a Low H. pylori Resistance Rate to IV-Related Compounds
3.4. Synergistic Interaction between Some IV-Related Compounds and Rabeprazole
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Brito, B.B.; Da Silva, F.A.F.; Soares, A.S.; Pereira, V.A.; Santos, M.L.C.; Sampaio, M.M.; Neves, P.H.M.; De Melo, F.F. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J. Gastroenterol. 2019, 25, 5578–5589. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.; Williams, D.W. Chapter 7–Helicobacter pylori. In Microbiology of Waterborne Diseases; Academic Press: New York, NY, USA, 2014; pp. 119–154. [Google Scholar]
- Zhang, X.-Y.; Zhang, P.-Y.; Aboul-Soud, M.A. From inflammation to gastric cancer: Role of Helicobacter pylori. Oncol. Lett. 2016, 13, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Neddermann, M.; Maubach, G.; Naumann, M. Pathogenesis of Helicobacter pylori infection. Helicobacter 2016, 21, 19–25. [Google Scholar] [CrossRef] [PubMed]
- WHO—IARC. Agents Classified by the IARC Monographs, Volumes 1–129. 2021. Available online: https://monographs.iarc.who.int/list-of-classifications (accessed on 11 July 2021).
- O’Connor, A.; Furuta, T.; Gisbert, J.P.; O’Morain, C. Review—Treatment of Helicobacter pylori infection 2020. Helicobacter 2020, 25, e12743. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Liou, J.; Lee, Y.; Wu, M. Treatment of Helicobacter pylori infection and its long-term impacts on gut microbiota. J. Gastroenterol. Hepatol. 2020, 35, 1107–1116. [Google Scholar] [CrossRef]
- Ghobadi, E.; Ghanbarimasir, Z.; Emami, S. A review on the structures and biological activities of anti-Helicobacter pylori agents. Eur. J. Med. Chem. 2021, 223, 113669. [Google Scholar] [CrossRef] [PubMed]
- Salillas, S.; Sancho, J. Flavodoxins as Novel Therapeutic Targets against Helicobacter pylori and Other Gastric Pathogens. Int. J. Mol. Sci. 2020, 21, 1881. [Google Scholar] [CrossRef] [PubMed]
- Cremades, N.; Bueno, M.; Toja, M.; Sancho, J. Towards a new therapeutic target: Helicobacter pylori flavodoxin. Biophys. Chem. 2005, 115, 267–276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freigang, J.; Diederichs, K.; Schäfer, K.P.; Welte, W.; Paul, R. Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori. Protein Sci. 2009, 11, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Cremades, N.; Velazquez-Campoy, A.; Freire, E.; Sancho, J. The Flavodoxin from Helicobacter pylori: Structural Determinants of Thermostability and FMN Cofactor Binding. Biochemistry 2007, 47, 627–639. [Google Scholar] [CrossRef][Green Version]
- Maurice, M.S.; Cremades, N.; Croxen, M.; Sisson, G.; Sancho, J.; Hoffman, P.S. Flavodoxin:Quinone Reductase (FqrB): A Redox Partner of Pyruvate:Ferredoxin Oxidoreductase That Reversibly Couples Pyruvate Oxidation to NADPH Production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 2007, 189, 4764–4773. [Google Scholar] [CrossRef]
- López-Llano, J.; Maldonado, S.; Bueno, M.; Lostao, A.; Jimenez, M.A.; Lillo, M.P.; Sancho, J. The Long and Short Flavodoxins: I. The role of the differentiating loop in apoflavodoxin structure and FMN binding. J. Biol. Chem. 2004, 279, 47177–47183. [Google Scholar] [CrossRef] [PubMed]
- Cremades, N.; Velázquez-Campoy, A.; Martínez-Júlvez, M.; Neira, J.L.; Pérez-Dorado, I.; Hermoso, J.; Jiménez, P.; Lanas, A.; Hoffman, P.S.; Sancho, J. Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents against Helicobacter pylori Infection. ACS Chem. Biol. 2009, 4, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Galano, J.J.; Alías, M.; Pérez, R.; Velázquez-Campoy, A.; Hoffman, P.S.; Sancho, J. Improved Flavodoxin Inhibitors with Potential Therapeutic Effects against Helicobacter pylori Infection. J. Med. Chem. 2013, 56, 6248–6258. [Google Scholar] [CrossRef]
- Salillas, S.; Alías, M.; Michel, V.; Mahía, A.; Lucía, A.; Rodrigues, L.; Bueno, J.; Galano-Frutos, J.J.; De Reuse, H.; Velázquez-Campoy, A.; et al. Design, Synthesis, and Efficacy Testing of Nitroethylene- and 7-Nitrobenzoxadiazol-Based Flavodoxin Inhibitors against Helicobacter pylori Drug-Resistant Clinical Strains and in Helicobacter pylori-Infected Mice. J. Med. Chem. 2019, 62, 6102–6115. [Google Scholar] [CrossRef] [PubMed]
- Flahou, B.; Haesebrouck, F.; Smet, A.; Yonezawa, H.; Osaki, T.; Kamiya, S. Gastric and Enterohepatic Non-Helicobacter pylori Helicobacters. Helicobacter 2013, 18, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Ménard, A.; Smet, A. Review: Other Helicobacter species. Helicobacter 2019, 24, e12645. [Google Scholar] [CrossRef] [PubMed]
- Smet, A.; Yahara, K.; Rossi, M.; Tay, A.; Backert, S.; Armin, E.; Fox, J.G.; Flahou, B.; Ducatelle, R.; Haesebrouck, F.; et al. Macroevolution of gastric Helicobacter species unveils interspecies admixture and time of divergence. ISME J. 2018, 12, 2518–2531. [Google Scholar] [CrossRef] [PubMed]
- Rahim, R.; Burrows, L.L.; Monteiro, M.A.; Perry, M.B.; Lam, J.S. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 2000, 146, 2803–2814. [Google Scholar] [CrossRef]
- Jacobs, M.A.; Alwood, A.; Thaipisuttikul, I.; Spencer, D.; Haugen, E.; Ernst, S.; Will, O.; Kaul, R.; Raymond, C.; Levy, R.; et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 14339–14344. [Google Scholar] [CrossRef]
- Held, K.; Ramage, E.; Jacobs, M.; Gallagher, L.; Manoil, C. Sequence-Verified Two-Allele Transposon Mutant Library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 2012, 194, 6387–6389. [Google Scholar] [CrossRef] [PubMed]
- McCallum, K.L.; Schoenhals, G.; Laakso, D.; Clarke, B.; Whitfield, C. A high-molecular-weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect. Immun. 1989, 57, 3816–3822. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Richards, J.C.; Perry, M.B.; Clarke, B.R.; MacLean, L.L. Expression of two structurally distinct D-galactan O antigens in the lipopolysaccharide of Klebsiella pneumoniae serotype O1. J. Bacteriol. 1991, 173, 1420–1431. [Google Scholar] [CrossRef]
- Clarke, B.R.; Whitfield, C. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: The rfb gene cluster is responsible for synthesis of the D-galactan I O polysaccharide. J. Bacteriol. 1992, 174, 4614–4621. [Google Scholar] [CrossRef]
- Avison, M.B.; Von Heldreich, C.J.; Higgins, C.S.; Bennett, P.M.; Walsh, T.R. A TEM-2 beta-lactamase encoded on an active Tn1-like transposon in the genome of a clinical isolate of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2000, 46, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Mamat, U.; Abda, E.M.; Kirchhoff, L.; Streit, W.; Schaible, U.E.; Niemann, S.; Kohl, T.A. Analysis of Phylogenetic Variation of Stenotrophomonas maltophilia Reveals Human-Specific Branches. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Van den Bulck, K.; Decostere, A.; Gruntar, I.; Baele, M.; Krt, B.; Ducatelle, R.; Haesebrouck, F. In Vitro Antimicrobial Susceptibility Testing of Helicobacter felis, H. bizzozeronii, and H. salomonis. Antimicrob. Agents Chemother. 2005, 49, 2997–3000. [Google Scholar] [CrossRef]
- Berlamont, H.; Smet, A.; De Bruyckere, S.; Boyen, F.; Ducatelle, R.; Haesebrouck, F.; De Witte, C. Antimicrobial susceptibility pattern of Helicobacter suis isolates from pigs and macaques. Vet. Microbiol. 2019, 239, 108459. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. M100-ED29, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Campos, L.A.; Sancho, J. Native-specific stabilization of flavodoxin by the FMN cofactor: Structural and thermodynamical explanation. Proteins Struct. Funct. Bioinform. 2006, 63, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, J.R.; Bloino, J.; Kudin, K.N.; Cossi, M.; Rega, N.; Pomelli, C.; et al. Gaussian 09, Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Morris, G.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Ramón-García, S.; Martín, C.; Aínsa, J.A.; De Rossi, E. Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 2006, 57, 252–259. [Google Scholar] [CrossRef]
- Aguilar-Pérez, C.; Gracia, B.; Rodrigues, L.; Vitoria, A.; Cebrian, R.; Brodin, P.; Maqueda, M.; Ainsa, J.; Song, O.-R. Synergy between circular bacteriocin AS-48 and ethambutol against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018, 62, e00359-18. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoints Tables for Interpretation of MICs and Zones Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 8 September 2021).
- Luo, H.; Lin, Y.; Gao, F.; Zhang, C.-T.; Zhang, R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements: Table 1. Nucleic Acids Res. 2013, 42, D574–D580. [Google Scholar] [CrossRef]
- Claveria-Gimeno, R.; Lanuza, P.M.; Morales-Chueca, I.; Torres, O.D.L.C.J.; Vega, S.; Abian, O.; Esteller, M.; Velazquez-Campoy, A. The intervening domain from MeCP2 enhances the DNA affinity of the methyl binding domain and provides an independent DNA interaction site. Sci. Rep. 2017, 7, srep41635. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wilson, T.J.M.; Jiang, Q.; Taylor, D.E. Spontaneous Mutations That Confer Antibiotic Resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2001, 45, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.; Gisbert, J.; Kuipers, E.; Axon, A.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.; et al. Management of Helicobacter pylori infection—the Maastricht V/Florence Consensus Report. Gut 2016, 66, 6–30. [Google Scholar] [CrossRef]
- Fallone, C.A.; Chiba, N.; van Zanten, S.V.; Fischbach, L.; Gisbert, J.P.; Hunt, R.H.; Jones, N.L.; Render, C.; Leontiadis, G.I.; Moayyedi, P.; et al. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology 2016, 151, 51–69.e14. [Google Scholar] [CrossRef]
- Flores-Treviño, S.; Mendoza-Olazarán, S.; Bocanegra-Ibarias, P.; Maldonado-Garza, H.J.; Garza-González, E. Helicobacter pylori drug resistance: Therapy changes and challenges. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Chey, W.D.; Leontiadis, G.I.; Howden, C.W.; Moss, S.F. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2017, 112, 212–239. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Molina-Infante, J.; Amador, J.; Bermejo, F.; Bujanda, L.; Calvet, X.; Castro-Fernandez, M.; Cuadrado-Lavín, A.; Elizalde, J.I.; Gene, E.; et al. IV Spanish consensus conference on Helicobacter pylori infection treatment. Gastroenterol. Hepatol. 2016, 39, 697–721. [Google Scholar] [CrossRef] [PubMed]
- Aboderin, O.A.; Abdu, A.R.; Odetoyin, B.; Okeke, I.N.; Lawal, O.; Ndububa, A.D.; Agbakwuru, E.A.; Lamikanra, A. Antibiotic resistance of Helicobacter pylori from patients in Ile-Ife, South-west, Nigeria. Afr. Health Sci. 2007, 7, 143–147. [Google Scholar] [CrossRef]
- Shriram, V.; Khare, T.; Bhagwat, R.; Shukla, R.; Kumar, V. Inhibiting Bacterial Drug Efflux Pumps via Phyto-Therapeutics to Combat Threatening Antimicrobial Resistance. Front. Microbiol. 2018, 9, 2990. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Ainsa, J.A.; Amaral, L.; Viveiros, M. Inhibition of drug efflux in mycobacteria with phenothiazines and other putative efflux inhibitors. Recent Pat. Anti-Infect. Drug Discov. 2011, 6, 118–127. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Masi, M.; Barbe, J. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 2005, 11, 382–389. [Google Scholar] [CrossRef]
- Debraekeleer, A.; Remaut, H. Future perspective for potential Helicobacter pylori eradication therapies. Future Microbiol. 2018, 13, 671–687. [Google Scholar] [CrossRef]
- Pule, C.M.; Sampson, S.L.; Warren, R.; Black, P.A.; Van Helden, P.D.; Victor, T.C.; Louw, G.E. Efflux pump inhibitors: Targeting mycobacterial efflux systems to enhance TB therapy. J. Antimicrob. Chemother. 2015, 71, 17–26. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Opperman, T.J.; Nguyen, S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015, 6, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Z.-Q.; Zheng, P.-Y.; Tang, F.-A.; Yang, P.-C. Influence of efflux pump inhibitors on the multidrug resistance of Helicobacter pylori. World J. Gastroenterol. 2010, 16, 1279–1284. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Villellas, C.; Bailo, R.; Viveiros, M.; Aínsa, J.A. Role of the Mmr Efflux Pump in Drug Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2012, 57, 751–757. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Amoako, D.G. Carbonyl Cyanide m-Chlorophenylhydrazine (CCCP) Reverses Resistance to Colistin, but Not to Carbapenems and Tigecycline in Multidrug-Resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef]
- Kumar, S.; Lekshmi, M.; Parvathi, A.; Ojha, M.; Wenzel, N.; Varela, M.F. Functional and Structural Roles of the Major Facilitator Superfamily Bacterial Multidrug Efflux Pumps. Microorganisms 2020, 8, 266. [Google Scholar] [CrossRef]
- Garvey, M.I.; Piddock, L.J.V. The Efflux Pump Inhibitor Reserpine Selects Multidrug-Resistant Streptococcus pneumoniae Strains That Overexpress the ABC Transporters PatA and PatB. Antimicrob. Agents Chemother. 2008, 52, 1677–1685. [Google Scholar] [CrossRef]
- Shaheen, A.; Afridi, W.A.; Mahboob, S.; Sana, M.; Zeeshan, N.; Ismat, F.; Mirza, O.; Iqbal, M.; Rahman, M. Reserpine Is the New Addition into the Repertoire of AcrB Efflux Pump Inhibitors. Mol. Microbiol. 2019, 53, 596–605. [Google Scholar] [CrossRef]
- Ramón-García, S.; Martin, C.; Thompson, C.J.; Aínsa, J.A. Role of the Mycobacterium tuberculosis P55 Efflux Pump in Intrinsic Drug Resistance, Oxidative Stress Responses, and Growth. Antimicrob. Agents Chemother. 2009, 53, 3675–3682. [Google Scholar] [CrossRef] [PubMed]
- Rindi, L. Efflux Pump Inhibitors Against Nontuberculous Mycobacteria. Int. J. Mol. Sci. 2020, 21, 4191. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, V.; Zullo, A.; Hassan, C.; Giorgio, F.; Rosania, R.; Ierardi, E. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World J. Gastrointest. Pathophysiol. 2011, 2, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Sharma, A.; Gupta, V.K. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. Genet. 2019, 17, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhu, F.; Ma, X.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspectives. Nat. Rev. Drug Discov. 2009, 8, 111–128. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Imamura, L.; PARK, J.; Kobashi, K. Helicobacter pylori Urease Inhibition by Rabeprazole, a Proton Pump Inhibitor. Biol. Pharm. Bull. 1995, 18, 1053–1056. [Google Scholar] [CrossRef] [PubMed]
- Langtry, H.D.; Markham, A. Rabeprazole: A review of its use in acid-related gastrointestinal disorders. Drugs 1999, 58, 725–742. [Google Scholar] [CrossRef]
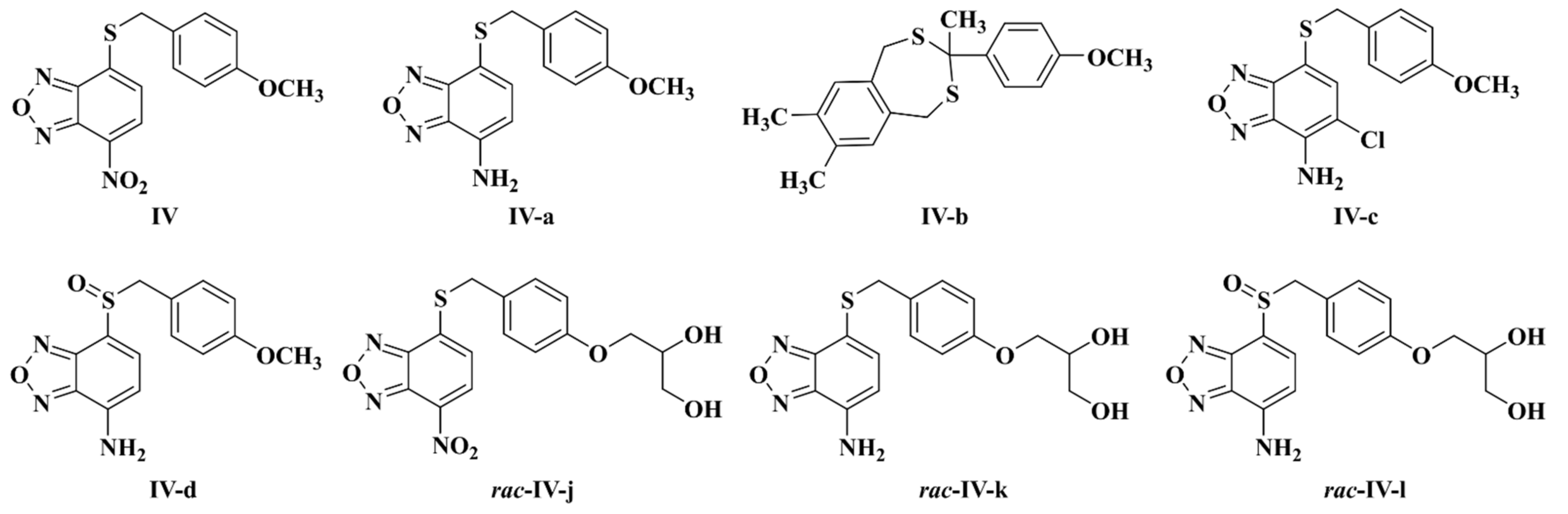



| Gram | Bacterial Species (Strain) | Compound MIC (μg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IV | IV-a | IV-b | IV-c | IV-d | rac- IV-j | rac- IV-k | rac-IV-l | Mnz | Cla | ||
| - | H. pylori (ATCC 700392) * | 2 | 8 | 1 | 2 | 8 | 1 | 16 | 256 | 2 | ≤0.032 |
| - | H. felis (JKM5) | 8 | 32 | ≤0.031 | |||||||
| - | H. suis (HS1) | 4 | 4 | 0.25 | |||||||
| (HS5) | 4 | 2 | ≤0.031 | ||||||||
| - | H. heilmannii (ASB1.4) | 2 | 2 | 0.125 | |||||||
| (ASB2) | 2 | 1 | ≤0.031 | ||||||||
| - | H. ailurogastricus (ASB7) | 4 | 1 | 0.125 | |||||||
| (ASB9) | 4 | 4 | 0.5 | ||||||||
| - | H. bizzozeronii (ASB22 kol15) | 16 | 16 | ≤0.031 | |||||||
| (10) | 8 | 32 | ≤0.031 | ||||||||
| (Heydar) | 8 | 32 | ≤0.031 | ||||||||
| - | H. hepaticus (ATCC 51449/3B1) | 64 | >64 | >64 | >64 | >64 | 64 | >64 | >64 | >64 | 0.063 |
| - | H. muridarum | 1 | >64 | >64 | >64 | >64 | 4 | >64 | >64 | >64 | >64 |
| - | H. bilis | 16 | >64 | >64 | >64 | >64 | 16 | >64 | >64 | >64 | 4 |
| - | C. jejuni (ATCC 33560) * | 2 | >64 | >64 | >64 | >64 | 4 | >64 | >64 | 1 | 4 |
| - | S. Typhimurium (SV 5015) * | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| - | E. coli (ATCC 10536) * | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 16 |
| - | P. aeruginosa (ATCC 15442) | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 8 |
| (MPAO1) | >512 | >512 | >512 | >512 | >512 | ||||||
| (PW9682) | >512 | >512 | >512 | >512 | >512 | ||||||
| - | S. maltophilia K279a | >512 | >512 | >512 | >512 | >512 | |||||
| (K279a ΔrmlBACD) | >512 | >512 | >512 | >512 | >512 | ||||||
| - | K. pneumoniae 3025 | >512 | >512 | >512 | >512 | >512 | |||||
| (CWK43) | >512 | >512 | >512 | >512 | >512 | ||||||
| + | Bacillus sp. (CECT 40) | 4 | >64 | >64 | >64 | >64 | 4 | >64 | >64 | >64 | 0.063 |
| + | S. pneumoniae (ATCC 49619) | 8 | >64 | >64 | 64 | >64 | 16 | >64 | >64 | >64 | ≤0.032 |
| + | L. monocytogenes (ATCCBAA-679) | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 0.25 |
| + | E. faecalis (JH2-2) | 2 | >64 | >64 | >64 | >64 | 4 | >64 | >64 | >64 | 0.25 |
| + | S. aureus (ATCC 29213) | 16 | >64 | >64 | >64 | >64 | 8 | >64 | >64 | >64 | ≤0.032 |
| BB pH 5 | 16 | 16–64 | 8–16 | ||||||||
| MH broth pH 7 | 16 | 16 | 0.5–1 | ||||||||
| + | C. diphtheriae (ATCC 39255) | 16 | 64 | >64 | 8 | >64 | 32 | >64 | >64 | >64 | 0.063 |
| + | C. ammoniagenes (ATCC 7862) | 16 | >64 | >64 | >64 | >64 | 16 | >64 | >64 | >64 | ≤0.032 |
| + | M. smegmatis (ATCC 700084) | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 4 |
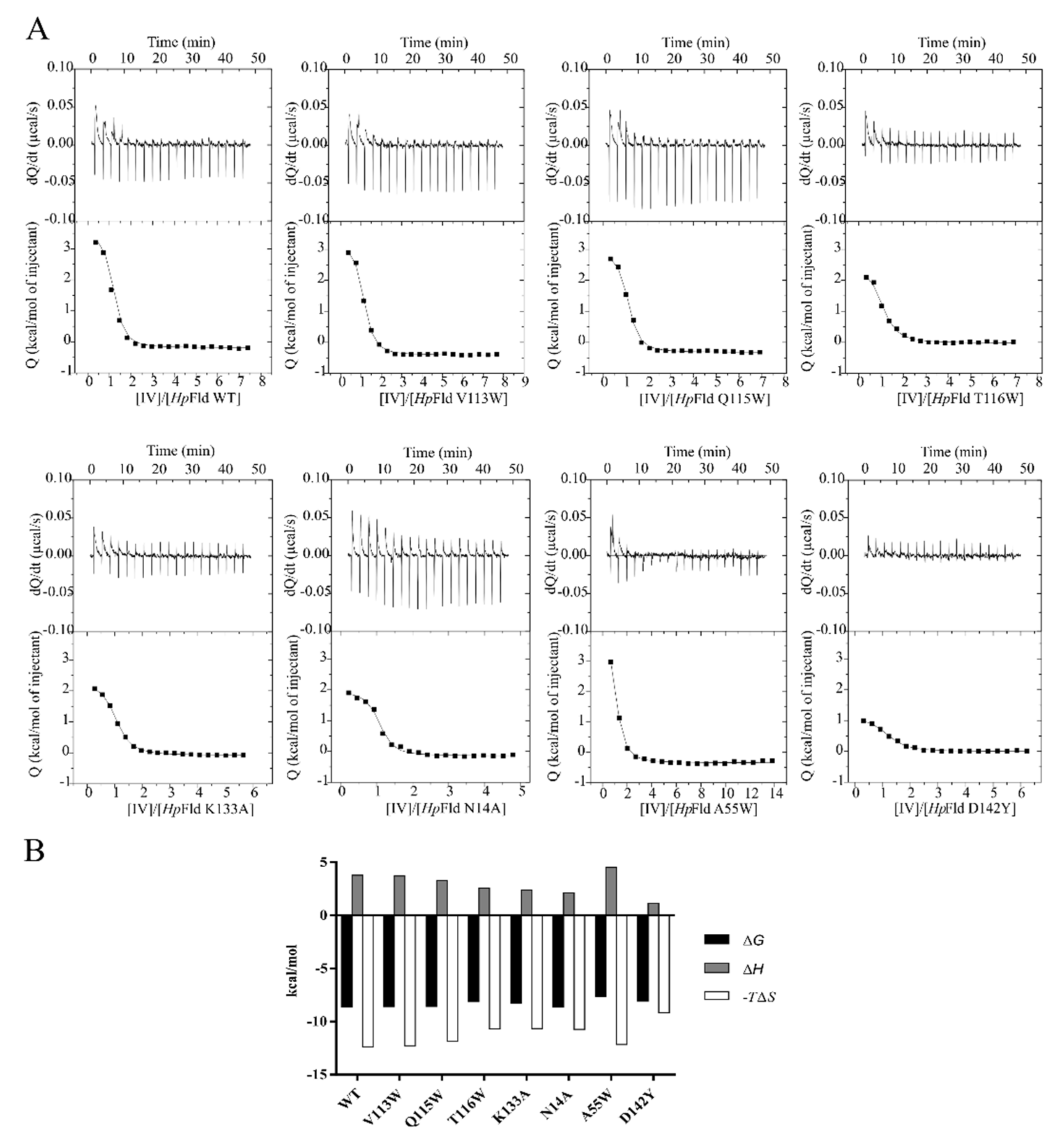
| Flavodoxin Variant | IV | IV-c | ||||||
|---|---|---|---|---|---|---|---|---|
| Kdb (μM) | ΔG c (kcal/mol) | ΔH d (kcal/mol) | −TΔS e (kcal/mol) | Kdb (μM) | ΔG c (kcal/mol) | ΔH d (kcal/mol) | −TΔS e (kcal/mol) | |
| WT | 0.48 | −8.58 | 3.77 | −12.35 | 0.67 | −8.39 | 3.10 | −11.49 |
| V113W | 0.50 | −8.56 | 3.69 | −12.25 | 1.45 | −7.93 | 1.40 | −9.33 |
| Q115W | 0.52 | −8.54 | 3.29 | −11.83 | 0.69 | −8.37 | 2.36 | −10.73 |
| T116W | 1.10 | −8.10 | 2.58 | −10.68 | 1.43 | −7.94 | 1.68 | −9.62 |
| K133A | 0.85 | −8.25 | 2.38 | −10.63 | 0.77 | −8.31 | 2.10 | −10.41 |
| N14A | 0.45 | −8.62 | 2.10 | −10.72 | 1.76 | −7.82 | 2.61 | −10.43 |
| A55W | 2.33 | −7.65 | 4.50 | −12.15 | 1.05 | −8.12 | 1.70 | −9.82 |
| D142Y | 1.18 | −8.05 | 1.13 | −9.18 | 0.44 | −8.64 | 0.92 | −9.56 |
| −8.29 ± 0.35 Mean ± SD | −8.19 ± 0.28 Mean ± SD | |||||||
| Compound | MIC a (μg/mL) | |||
|---|---|---|---|---|
| Without any EI | CCCP | Reserpine | Valinomycin | |
| IV | 2 | 0.5 | 0.5 | 0.5 |
| IV-a | 8 | 8 | 4 | 4 |
| IV-b | 1 | 2 | 2 | 2 |
| IV-c | 2 | 4 | 4 | 4 |
| IV-d | 8 | 32 | 32 | 32 |
| rac-IV-j | 1 | 4 | 2 | 2 |
| rac-IV-k | 16 | 16 | 16 | 8 |
| rac-IV-l | >64 | >64 | >64 | >64 |
| Mnz | 2 | 4 | 4 | 2 |
| Cla | ≤0.032 | ≤0.032 | ≤0.032 | ≤0.032 |
| Combination Compound | IV-Related Compound | FICcombination compound | FICIV-related compound | FICI b |
|---|---|---|---|---|
| IV-a | 0.016 | 0.5 | 0.516 | |
| IV-b | 1 | 1 | 2 | |
| IV-c | 1 | 1 | 2 | |
| IV | IV-d | 1 | 1 | 2 |
| rac-IV-j | 0.25 | 0.5 | 0.75 | |
| rac-IV-k | 1 | 1 | 2 | |
| rac-IV-l | 0.25 | 0.5 | 0.75 | |
| IV | 0.063 | 0.5 | 0.56 | |
| IV-a | 0.5 | 0.25 | 0.75 | |
| IV-b | 0.5 | 0.5 | 1 | |
| Mnz | IV-c | 0.5 | 0.5 | 1 |
| IV-d | 0.5 | 0.5 | 1 | |
| rac-IV-j | 0.5 | 0.5 | 1 | |
| rac-IV-k | 0.5 | 0.5 | 1 | |
| rac-IV-l | 0.5 | 0.5 | 1 | |
| IV | 0.5 | 0.5 | 1 | |
| IV-a | 0.5 | 0.25 | 0.75 | |
| IV-b | 0.5 | 0.5 | 1 | |
| Cla | IV-c | 0.5 | 0.25 | 0.75 |
| IV-d | 0.5 | 0.125 | 0.62 | |
| rac-IV-j | 0.5 | 0.125 | 0.62 | |
| rac-IV-k | 0.25 | 0.5 | 0.75 | |
| rac-IV-l | 0.5 | 0.125 | 0.62 | |
| IV | 0.5 | 0.5 | 1 | |
| IV-a | 0.25 | 0.5 | 0.75 | |
| IV-b | 0.125 | 0.5 | 0.62 | |
| Omeprazole | IV-d | 0.25 | 0.5 | 0.75 |
| rac-IV-j | 0.5 | 0.5 | 1 | |
| rac-IV-k | 0.25 | 0.5 | 0.75 | |
| rac-IV-l | 0.5 | 0.5 | 1 | |
| IV | 1 | 1 | 2 | |
| IV-a | 0.5 | 0.063 | 0.56 | |
| IV-b | 0.25 | 0.25 | 0.5 | |
| Rabeprazole | IV-d | 0.5 | 0.125 | 0.62 |
| rac-IV-j | 0.5 | 0.5 | 1 | |
| rac-IV-k | 0.5 | 0.5 | 1 | |
| rac-IV-l | 0.25 | 0.25 | 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salillas, S.; Galano-Frutos, J.J.; Mahía, A.; Maity, R.; Conde-Giménez, M.; Anoz-Carbonell, E.; Berlamont, H.; Velazquez-Campoy, A.; Touati, E.; Mamat, U.; et al. Selective Targeting of Human and Animal Pathogens of the Helicobacter Genus by Flavodoxin Inhibitors: Efficacy, Synergy, Resistance and Mechanistic Studies. Int. J. Mol. Sci. 2021, 22, 10137. https://doi.org/10.3390/ijms221810137
Salillas S, Galano-Frutos JJ, Mahía A, Maity R, Conde-Giménez M, Anoz-Carbonell E, Berlamont H, Velazquez-Campoy A, Touati E, Mamat U, et al. Selective Targeting of Human and Animal Pathogens of the Helicobacter Genus by Flavodoxin Inhibitors: Efficacy, Synergy, Resistance and Mechanistic Studies. International Journal of Molecular Sciences. 2021; 22(18):10137. https://doi.org/10.3390/ijms221810137
Chicago/Turabian StyleSalillas, Sandra, Juan José Galano-Frutos, Alejandro Mahía, Ritwik Maity, María Conde-Giménez, Ernesto Anoz-Carbonell, Helena Berlamont, Adrian Velazquez-Campoy, Eliette Touati, Uwe Mamat, and et al. 2021. "Selective Targeting of Human and Animal Pathogens of the Helicobacter Genus by Flavodoxin Inhibitors: Efficacy, Synergy, Resistance and Mechanistic Studies" International Journal of Molecular Sciences 22, no. 18: 10137. https://doi.org/10.3390/ijms221810137
APA StyleSalillas, S., Galano-Frutos, J. J., Mahía, A., Maity, R., Conde-Giménez, M., Anoz-Carbonell, E., Berlamont, H., Velazquez-Campoy, A., Touati, E., Mamat, U., Schaible, U. E., Gálvez, J. A., Díaz-de-Villegas, M. D., Haesebrouck, F., Aínsa, J. A., & Sancho, J. (2021). Selective Targeting of Human and Animal Pathogens of the Helicobacter Genus by Flavodoxin Inhibitors: Efficacy, Synergy, Resistance and Mechanistic Studies. International Journal of Molecular Sciences, 22(18), 10137. https://doi.org/10.3390/ijms221810137








