Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs
Abstract
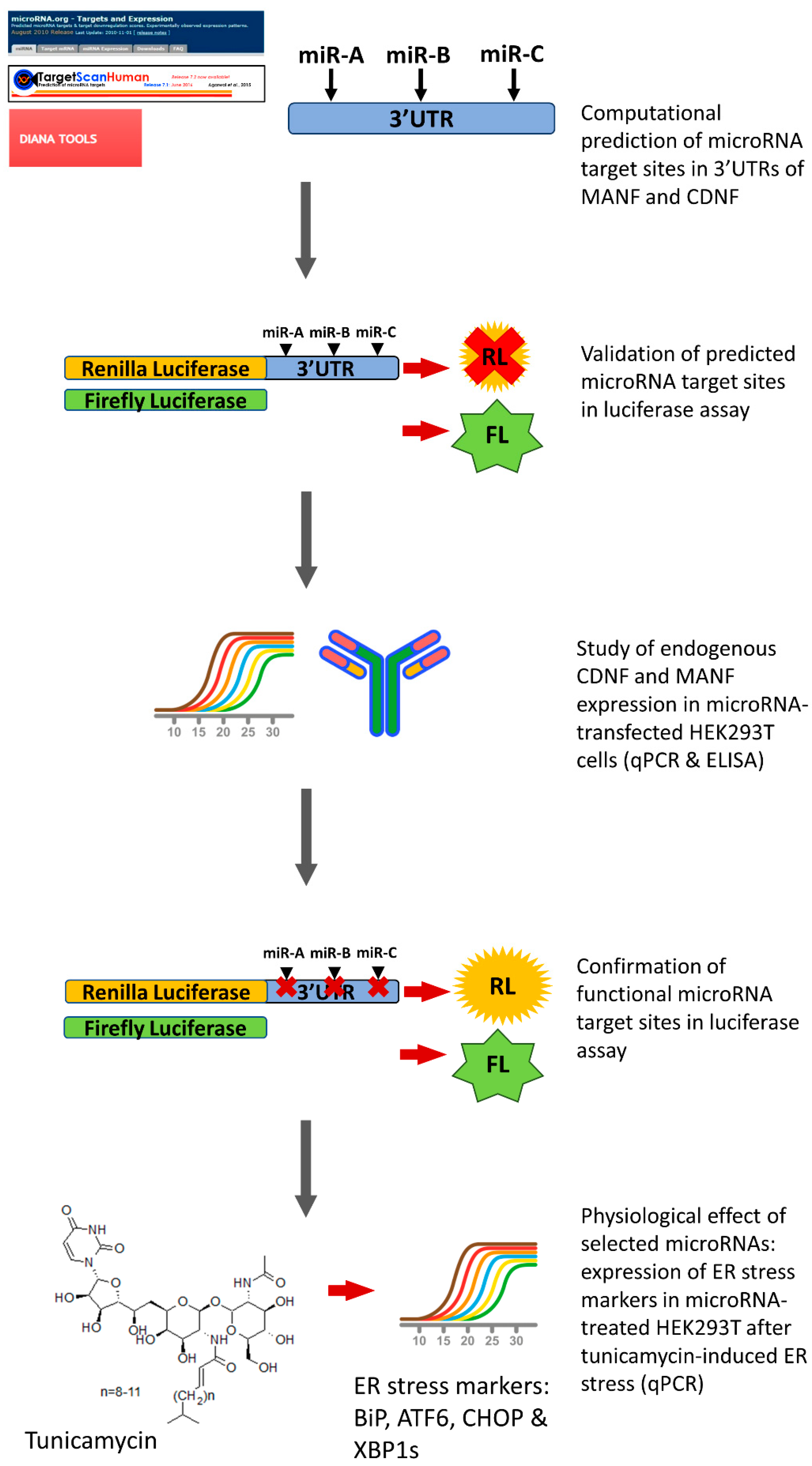
:1. Introduction
2. Results
2.1. miRNAs, Predicted to Bind 3′UTR of Human MANF and CDNF
2.2. miRNA Regulation of Protein Synthesis from Transcripts Containing hMANF and hCDNF 3′UTRs
2.3. Regulation of Endogenous hMANF and hCDNF by miRNAs
2.4. Confirmation of miRNAs’ Predicted Binding Sites
2.5. Physiological Effect of miRNAs, Targeting Expression of Human MANF and CDNF
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Prediction of miRNAs, Targeting hMANF and hCDNF
4.2. Cell Culture
4.3. Plasmids
4.4. Site-Directed Mutagenesis
4.5. Dual-Luciferase Reporter Assay
4.6. Quantitative Real-Time PCR (RT-qPCR)
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindholm, P.; Voutilainen, M.; Laurén, J.; Peränen, J.; Leppänen, V.-M.; Andressoo, J.-O.; Lindahl, M.; Janhunen, S.; Kalkkinen, N.; Timmusk, T.; et al. Novel neurotrophic factor CDNF protects and rescues midbrain dopamine neurons in vivo. Nat. Cell Biol. 2007, 448, 73–77. [Google Scholar] [CrossRef]
- Jӓntti, M.; Harvey, B.K. Trophic activities of endoplasmic reticulum proteins CDNF and MANF. Cell Tissue Res. 2020, 382, 83–100. [Google Scholar] [CrossRef]
- Lindholm, P.; Saarma, M. Novel CDNF/MANF family of neurotrophic factors. Dev. Neurobiol. 2010, 70, 360–371. [Google Scholar] [CrossRef]
- Montaser, H.; Patel, K.A.; Balboa, D.; Ibrahim, H.; Lithovius, V.; Näätänen, A.; Chandra, V.; Demir, K.; Acar, S.; Ben-Omran, T.; et al. Loss of MANF Causes Childhood-Onset Syndromic Diabetes Due to Increased Endoplasmic Reticulum Stress. Diabetes 2021, 70, 1006–1018. [Google Scholar] [CrossRef]
- Friedman, R.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Gerasymchuk, D.; Hubiernatorova, A.; Domanskyi, A. MicroRNAs Regulating Cytoskeleton Dynamics, Endocytosis, and Cell Motility—A Link Between Neurodegeneration and Cancer? Front. Neurol. 2020, 11, 549006. [Google Scholar] [CrossRef]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef] [Green Version]
- Emde, A. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.B.; Uehara, H.; Kim, Y.; Yasuma, T.; Yasuma, R.; Hirahara, S.; Makin, R.D.; Apicella, I.; Pereira, F.; Nagasaka, Y.; et al. Chronic Dicer1 deficiency promotes atrophic and neovascular outer retinal pathologies in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 2579–2587. [Google Scholar] [CrossRef] [Green Version]
- Juźwik, C.A.; Drake, S.S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E. MicroRNA alterations in iPSC-derived dopaminergic neurons from Parkinson disease patients. Neurobiol. Aging 2018, 69, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5–23. [Google Scholar] [CrossRef]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef] [Green Version]
- Varendi, K.; Kumar, A.; Härma, M.-A.; Andressoo, J.-O. miR-1, miR-10b, miR-155, and miR-191 are novel regulators of BDNF. Cell. Mol. Life Sci. 2014, 71, 4443–4456. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.T. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012, 72, 269–277. [Google Scholar] [CrossRef]
- Maheu, M.; Lopez, J.P.; Crapper, L.; Davoli, M.A.; Turecki, G.; Mechawar, N. MicroRNA regulation of central glial cell line-derived neurotrophic factor (GDNF) signalling in depression. Transl. Psychiatry 2015, 5, e511. [Google Scholar] [CrossRef] [Green Version]
- Coutinho de Almeida, R. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann. Rheum. Dis. 2019, 78, 270–277. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.-J.; Yuan, Y.-G.; Das, J.; Yasuda, H.; Kim, J.-H. MicroRNA-7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Georges, S.A.; Biery, M.C.; Kim, S.-Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated Regulation of Cell Cycle Transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008, 68, 10105–10112. [Google Scholar] [CrossRef] [Green Version]
- Hollander, J.A.; Im, H.-I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microRNA controls cocaine intake through CREB signalling. Nat. Cell Biol. 2010, 466, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef]
- Wu, J.; Chen, S.; Liu, H.; Zhang, Z.; Ni, Z.; Chen, J.; Yang, Z.; Nie, Y.; Fan, D. Tunicamycin specifically aggravates ER stress and overcomes chemoresistance in multidrug-resistant gastric cancer cells by inhibiting N-glycosylation. J. Exp. Clin. Cancer Res. 2018, 37, 272. [Google Scholar] [CrossRef]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef]
- Pakarinen, E.; Danilova, T.; Voikar, V.; Chmielarz, P.; Piepponen, P.; Airavaara, M.; Saarma, M.; Lindahl, M. MANF Ablation Causes Prolonged Activation of the UPR without Neurodegeneration in the Mouse Midbrain Dopamine System. eNeuro 2020, 7. [Google Scholar] [CrossRef]
- Tseng, K.-Y.; Danilova, T.; Domanskyi, A.; Saarma, M.; Lindahl, M.; Airavaara, M. MANF Is Essential for Neurite Extension and Neuronal Migration in the Developing Cortex. eNeuro 2017, 4. [Google Scholar] [CrossRef]
- Xu, S. Mesencephalic astrocyte-derived neurotrophic factor (MANF) protects against Abeta toxicity via attenuating Abeta-induced endoplasmic reticulum stress. J. Neuroinflam. 2019, 16, 35. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, M. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep. 2014, 7, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W. Cerebral dopamine neurotrophic factor alleviates Abeta25-35-induced endoplasmic reticulum stress and early synapto-toxicity in rat hippocampal cells. Neurosci. Lett. 2016, 633, 40–46. [Google Scholar] [CrossRef]
- Lindahl, M. Cerebral dopamine neurotrophic factor-deficiency leads to degeneration of enteric neurons and altered brain dopa-mine neuronal function in mice. Neurobiol. Dis. 2020, 134, 104696. [Google Scholar] [CrossRef]
- Yavarna, T.; Al-Dewik, N.; Al-Mureikhi, M.; Ali, R.; Al-Mesaifri, F.; Mahmoud, L.; Shahbeck, N.; Lakhani, S.; AlMulla, M.; Nawaz, Z.; et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Qual. Life Res. 2015, 134, 967–980. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, F.; Yang, Q.; Zhang, Y.; Liu, Q.; Jiang, W.; Cao, H.; Li, D.; Xie, S.; Tong, N.; et al. Circulating mesencephalic astrocyte-derived neurotrophic factor is increased in newly diagnosed prediabetic and diabetic patients, and is associated with insulin resistance. Endocr. J. 2017, 64, 403–410. [Google Scholar] [CrossRef] [Green Version]
- Chen, L. Mesencephalic astrocyte-derived neurotrophic factor is involved in inflammation by negatively regulating the NF-kappaB pathway. Sci. Rep. 2015, 5, 8133. [Google Scholar] [CrossRef]
- Galli, E.; Planken, A.; Kadastik-Eerme, L.; Saarma, M.; Taba, P.; Lindholm, P. Increased Serum Levels of Mesencephalic Astrocyte-Derived Neurotrophic Factor in Subjects With Parkinson’s Disease. Front. Neurosci. 2019, 13, 929. [Google Scholar] [CrossRef] [Green Version]
- Joshi, H.; McIntyre, W.B.; Kooner, S.; Rathbone, M.; Gabriele, S.; Gabriele, J.; Baranowski, D.; Frey, B.N.; Mishra, R.K. Decreased Expression of Cerebral Dopamine Neurotrophic Factor in Platelets of Stroke Patients. J. Stroke Cerebrovasc. Dis. 2019, 29, 104502. [Google Scholar] [CrossRef] [Green Version]
- Virachit, S.; Mathews, K.J.; Cottam, V.; Werry, E.; Galli, E.; Rappou, E.; Lindholm, P.; Saarma, M.; Halliday, G.M.; Weickert, C.S.; et al. Levels of glial cell line-derived neurotrophic factor are decreased, but fibroblast growth factor 2 and cerebral dopamine neurotrophic factor are increased in the hippocampus in Parkinson’s disease. Brain Pathol. 2019, 29, 813–825. [Google Scholar] [CrossRef]
- Inukai, S.; Slack, F. MicroRNAs and the Genetic Network in Aging. J. Mol. Biol. 2013, 425, 3601–3608. [Google Scholar] [CrossRef] [Green Version]
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, P.; Erfle, H.; Sonntag, K.-C.; Schütz, G.; Vinnikov, I.A.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, e2813. [Google Scholar] [CrossRef] [Green Version]
- Simunovic, F.; Yi, M.; Wang, Y.; Stephens, R.; Sonntag, K.C. Evidence for Gender-Specific Transcriptional Profiles of Nigral Dopamine Neurons in Parkinson Disease. PLoS ONE 2010, 5, e8856. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, G.T. A cross-study transcriptional analysis of Parkinson’s disease. PLoS ONE 2009, 4, e4955. [Google Scholar] [CrossRef] [Green Version]
- Santos-Lobato, B.L.; Vidal, A.F.; Ribeiro-Dos-Santos, A. Regulatory miRNA-mRNA Networks in Parkinson’s Disease. Cells 2021, 10, 1410. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Tatura, R.; Kraus, T.; Giese, A.; Arzberger, T.; Buchholz, M.; Höglinger, G.; Müller, U. Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Park. Relat. Disord. 2016, 33, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Persengiev, S.; Kondova, I.; Otting, N.; Koeppen, A.H.; Bontrop, R. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 2011, 32, 2316.e17–2316.e27. [Google Scholar] [CrossRef]
- Moradifard, S.; Hoseinbeyki, M.; Ganji, S.M.; Minuchehr, Z. Analysis of microRNA and Gene Expression Profiles in Alzheimer’s Disease: A Meta-Analysis Approach. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Tsivinsky, V.G.; Crawford, F.; Mullan, M.; Abdullah, L.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging 2012, 4, 590–605. [Google Scholar] [CrossRef] [Green Version]
- Sheinerman, K.S.; Tsivinsky, V.G.; Abdullah, L.; Crawford, F.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment: Biomarker Validation Study. Aging 2013, 5, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Karolina, D.S. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Maurel, M.; Chevet, E. Endoplasmic reticulum stress signaling: The microRNA connection. Am. J. Physiol. Physiol. 2013, 304, C1117–C1126. [Google Scholar] [CrossRef] [Green Version]
- Leung, A.K.; Calabrese, J.M.; Sharp, P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA 2006, 103, 18125–18130. [Google Scholar] [CrossRef] [Green Version]
- Linsen, S.E.; Tops, B.B.; Cuppen, E. miRNAs: Small changes, widespread effects. Cell Res. 2008, 18, 1157–1159. [Google Scholar] [CrossRef] [Green Version]
- Narasimhan, M.; Patel, D.; Vedpathak, D.; Rathinam, M.; Henderson, G.; Mahimainathan, L. Identification of Novel microRNAs in Post-Transcriptional Control of Nrf2 Expression and Redox Homeostasis in Neuronal, SH-SY5Y Cells. PLoS ONE 2012, 7, e51111. [Google Scholar] [CrossRef] [Green Version]
- Digaleh, H.; Kiaei, M.; Khodagholi, F. Nrf2 and Nrf1 signaling and ER stress crosstalk: Implication for proteasomal degradation and autophagy. Cell. Mol. Life Sci. 2013, 70, 4681–4694. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, H.; Zhu, D.; Huang, H.; Liu, G.; Lun, P. Targeted Suppression of Chaperone-Mediated Autophagy by miR-320a Promotes α-Synuclein Aggregation. Int. J. Mol. Sci. 2014, 15, 15845–15857. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, C.; Gu, C.; Cui, X.; Wu, J. MiRNA-144-3p inhibits high glucose induced cell proliferation through suppressing FGF16. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Tseng, K.-Y.; Anttila, J.E.; Khodosevich, K.; Tuominen, R.K.; Lindahl, M.; Domanskyi, A.; Airavaara, M. MANF Promotes Differentiation and Migration of Neural Progenitor Cells with Potential Neural Regenerative Effects in Stroke. Mol. Ther. 2018, 26, 238–255. [Google Scholar] [CrossRef] [Green Version]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Gentner, B.; Schira, G.; Giustacchini, A.; Amendola, M.; Brown, B.D.; Ponzoni, M.; Naldini, L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Chem. Biol. 2008, 6, 63–66. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Knauss, J.L.; Bian, S.; Sun, T. Plasmid-based target protectors allow specific blockade of miRNA silencing activity in mammalian de-velopmental systems. Front. Cell Neurosci. 2013, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Galli, E.; Lindholm, P.; Kontturi, L.-S.; Saarma, M.; Urtti, A.; Yliperttula, M. Characterization of CDNF-Secreting ARPE-19 Cell Clones for Encapsulated Cell Therapy. Cell Transplant. 2019, 28, 413–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galli, E. Increased circulating concentrations of mesencephalic astrocyte-derived neurotrophic factor in children with type 1 diabetes. Sci. Rep. 2016, 6, 29058. [Google Scholar] [CrossRef]
- Lew, M. Good statistical practice in pharmacology Problem 2. Br. J. Pharmacol. 2007, 152, 299–303. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konovalova, J.; Gerasymchuk, D.; Arroyo, S.N.; Kluske, S.; Mastroianni, F.; Pereyra, A.V.; Domanskyi, A. Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs. Int. J. Mol. Sci. 2021, 22, 9691. https://doi.org/10.3390/ijms22189691
Konovalova J, Gerasymchuk D, Arroyo SN, Kluske S, Mastroianni F, Pereyra AV, Domanskyi A. Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs. International Journal of Molecular Sciences. 2021; 22(18):9691. https://doi.org/10.3390/ijms22189691
Chicago/Turabian StyleKonovalova, Julia, Dmytro Gerasymchuk, Sergio Navarette Arroyo, Sven Kluske, Francesca Mastroianni, Alba Vargas Pereyra, and Andrii Domanskyi. 2021. "Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs" International Journal of Molecular Sciences 22, no. 18: 9691. https://doi.org/10.3390/ijms22189691
APA StyleKonovalova, J., Gerasymchuk, D., Arroyo, S. N., Kluske, S., Mastroianni, F., Pereyra, A. V., & Domanskyi, A. (2021). Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs. International Journal of Molecular Sciences, 22(18), 9691. https://doi.org/10.3390/ijms22189691







