Generation of a Triadin KnockOut Syndrome Zebrafish Model
Abstract
1. Introduction
2. Results
2.1. Zebrafish Trdn Identification and Spatio-Temporal Expression Analyses
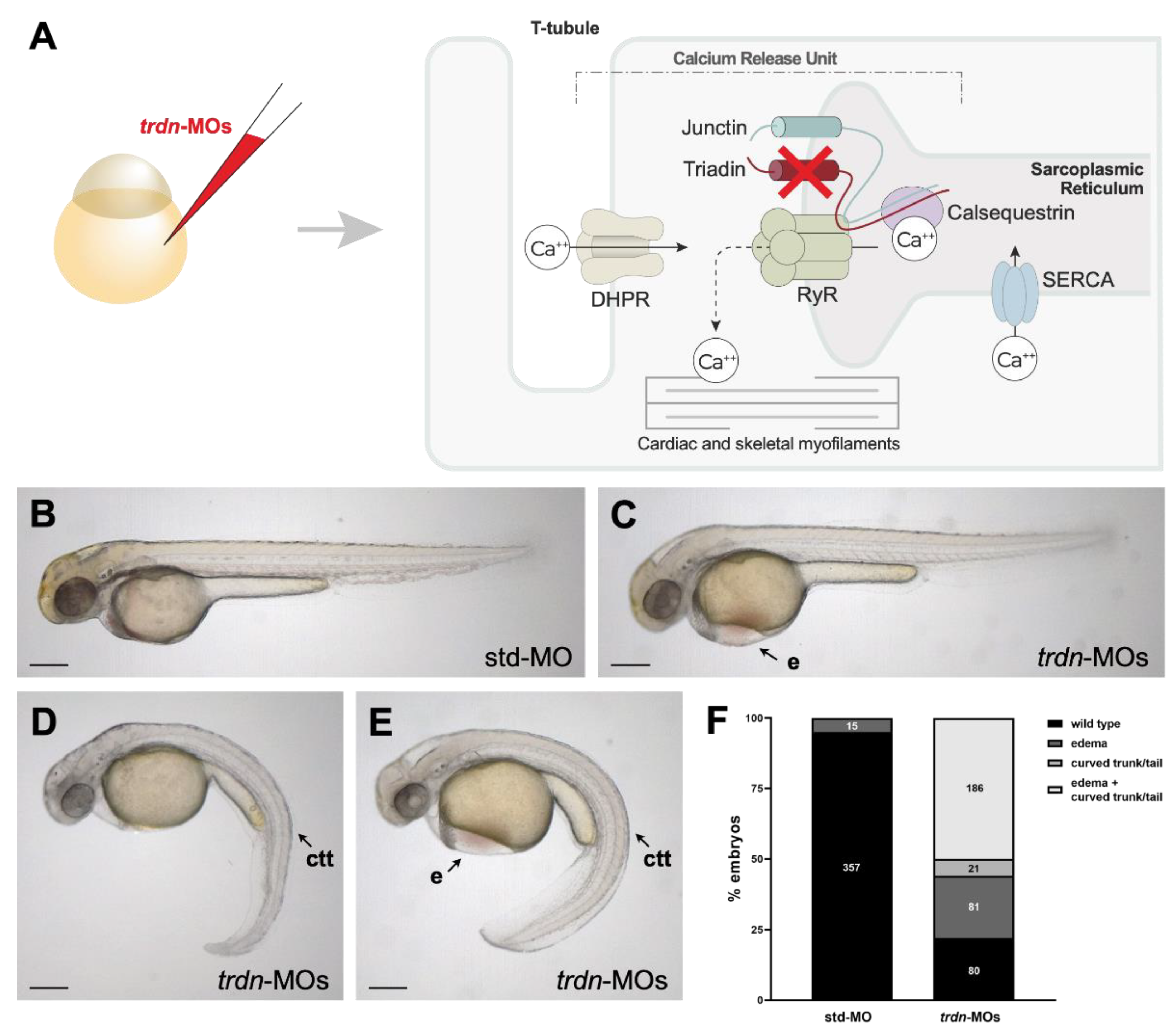
2.2. Generation and Characterization of a Trdn Loss-of-Function Model in Zebrafish
2.3. The General Morphology and Organization of Skeletal Muscle Fibres Are Not Impaired by Trdn Knockdown
2.4. trdn Loss-of-Function Impaired the Heart Rhythm
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Microinjection
4.3. RT-PCR
4.4. Western Blot
4.5. Imaging
4.6. Whole-Mount In Situ Hybridization (WISH)
4.7. Immunofluorescence
4.8. Semithin Sections
4.9. Heart Rhythm Detection
4.10. Drug Treatment
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahra, R. Global public health problem of sudden cardiac death. J. Electrocardiol. 2007, 40, S118–S122. [Google Scholar] [CrossRef]
- Sana, M.A.K.; William, G.S.; Michael, J.A.; William, J.B.; David, J.C.; Anne, B.C.; Barbara, J.D.; Timm, D.; Michael, E.F.; Gregg, C.F.; et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: Executive summary: A report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines and the heart rhythm society. Circulation 2018, 138, e210–e271. [Google Scholar] [CrossRef]
- Scrocco, C.; Bezzina, C.R.; Ackerman, M.J.; Brhr, E.R. Genetics and genomics of arrhythmic risk: Current and future strategies to prevent sudden cardiac death. Nat. Rev. Cardiol. 2021, 1–11. [Google Scholar] [CrossRef]
- Schwartz, P.J.; Ackerman, M.J.; Antzelevitch, C.; Bezzina, C.R.; Borggrefe, M.; Cuneo, B.F.; Wilde, A.A.M. Inherited cardiac arrhythmias. Nat. Rev. Dis. Primers 2020, 6, 1–22. [Google Scholar] [CrossRef]
- Altmann, H.M.; Tester, D.J.; Will, M.L.; Middha, S.; Evans, J.M.; Eckloff, B.W.; Ackerman, M.J. Homozygous/Compound heterozygous triadin mutations associated with autosomal-recessive long-qt syndrome and pediatric sudden cardiac arrest: Elucidation of the triadin knockout syndrome. Circulation 2015, 131, 2051–2060. [Google Scholar] [CrossRef]
- Roux-Buisson, N.; Cacheux, M.; Fourest-Lieuvin, A.; Fauconnier, J.; Brocard, J.; Denjoy, I.; Durand, P.; Guicheney, P.; Kyndt, F.; Leenhardt, A. Absence of triadin, a protein of the calcium release complex, is responsible for cardiac arrhythmia with sudden death in human. Hum. Mol. Genet. 2012, 21, 2759–2767. [Google Scholar] [CrossRef]
- Rooryck, C.; Kyndt, F.; Bozon, D.; Roux-Buisson, N.; Sacher, F.; Probst, V.; Thambo, J.B. New family with catecholaminergic polymorphic ventricular tachycardia linked to the triadin gene. J. Cardiovasc. Electrophysiol. 2015, 26, 1146–1150. [Google Scholar] [CrossRef]
- Walsh, M.A.; Stuart, A.G.; Schlecht, H.B.; James, A.F.; Hancox, J.C.; Newbury-Ecob, R.A. Compound heterozygous triadin mutation causing cardiac arrest in two siblings. Pacing Clin. Electrophysiol. PACE 2016, 39, 497–501. [Google Scholar] [CrossRef]
- O’Callaghan, B.M.; Hancox, J.C.; Stuart, A.G.; Armstrong, C.; Williams, M.M.; Hills, A.; Pearce, H.; Dent, C.L.; Gable, M.; Walsh, M.A. A unique triadin exon deletion causing a null phenotype. HeartRhythm Case Rep. 2018, 4, 514–518. [Google Scholar] [CrossRef]
- Clemens, D.J.; Tester, D.J.; Giudicessi, J.R.; Bos, J.M. International triadin knockout syndrome registry. Circ. Genom. Precis. Med. 2019, 12, 65–75. [Google Scholar] [CrossRef]
- Clemens, D.J.; Tester, D.J.; Marty, I.; Ackerman, M.J. Phenotype-guided whole genome analysis in a patient with genetically elusive long QT syndrome yields a novel TRDN-encoded triadin pathogenetic substrate for triadin knockout syndrome and reveals a novel primate-specific cardiac TRDN transcript. Heart Rhythm 2020, 17, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Gigli, L.; Gamberucci, A.; Bordoni, R.; Resta, C.D. A novel homozygous mutation in the TRDN gene causes a severe form of pediatric malignant ventricular arrhythmia. Heart Rhythm 2020, 17, 296–304. [Google Scholar] [CrossRef]
- Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Cesar, S.; Arbelo, E.; Jorda, P.; García-Álvarez, A.; Cruzalegui, J.C.; Merchan, E.F.; Fiol, V.; Brugada, J.; et al. Pediatric malignant arrhythmias caused by rare homozygous genetic variants in TRDN: A comprehensive interpretation. Front. Pediatrics 2021, 8, 601708. [Google Scholar] [CrossRef]
- Marty, I. Triadin regulation of the ryanodine receptor complex. J. Physiol. 2015, 593, 3261–3266. [Google Scholar] [CrossRef]
- Thevenon, D.; Smida-Rezgui, S.; Chevessier, F.; Groh, S.; Henry-Berger, J.; Romero, N.B.; Villaz, M.; DeWaard, M.; Marty, I. Human skeletal muscle triadin: Gene organization and cloning of the major isoform, Trisk 51. Biochem. Biophys. Res. Commun. 2003, 303, 669–675. [Google Scholar] [CrossRef]
- Vassilopoulos, S.; Thevenon, D.; Rezgui, S.S.; Brocard, J.; Chapel, A.; Lacampagne, A.; Lunardi, J.; Dewaard, M.; Marty, I. Triadins are not triad-specific proteins: Two new skeletal muscle triadins possibly involved in the architecture of sarcoplasmic reticulum. J. Biol. Chem. 2005, 280, 28601–28609. [Google Scholar] [CrossRef]
- Kobayashi, Y.M.; Jones, L.R. Identification of triadin 1 as the predominant triadin isoform expressed in mammalian myocardium. J. Biol. Chem. 1999, 274, 28660–28668. [Google Scholar] [CrossRef][Green Version]
- Barone, V.; Randazzo, D.; Delre, V.; Sorrentino, V.; Rossi, D. Organization of junctional sarcoplasmic reticulum proteins in skeletal muscle fibers. J. Muscle Res. Cell Motil. 2015, 36, 501–515. [Google Scholar] [CrossRef]
- Zhang, L.; Kelley, J.; Schmeisser, G.; Kobayashi, Y.M.; Jones, L.R. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 1997, 272, 23389–23397. [Google Scholar] [CrossRef]
- Chopra, N.; Yang, T.; Asghari, P.; Moore, E.D.; Huke, S.; Akin, B.; Cattolica, R.A.; Perez, C.F.; Hlaing, T.; Knollmann-Ritschel, B.E. Ablation of triadin causes loss of cardiac Ca2+ release units, impaired excitation-contraction coupling, and cardiac arrhythmias. Proc. Natl. Acad. Sci. USA 2009, 106, 7636–7641. [Google Scholar] [CrossRef]
- Oddoux, S.; Brocard, J.; Schweitzer, A.; Szentesi, P.; Giannesini, B.; Brocard, J.; Faure, J.; Pernet-Gallay, K.; Bendahan, D.; Lunardi, J.; et al. Triadin deletion induces impaired skeletal muscle function. J. Biol. Chem. 2009, 284, 34918–34920. [Google Scholar] [CrossRef]
- Rabbani, B.; Khorgami, M.; Dalili, M.; Zamani, N.; Mahdieh, N.; Gollob, M.H. Novel cases of pediatric sudden cardiac death secondary to TRDN mutations presenting as long QT syndrome at rest and catecholaminergic polymorphic ventricular tachycardia during exercise: The TRDN arrhythmia syndrome. Am. J. Med. Genet. Part A 2021. [Google Scholar] [CrossRef]
- Yalcin, H.C.; Amindari, A.; Butcher, J.T.; Althani, A.; Yacoub, M. Heart function and hemodynamic analysis for zebrafish embryos. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2017, 246, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Stellabotte, F.; Dobbs-Mcauliffe, B.; Fernandez, D.A.; Feng, X.; Devoto, S.H. Dynamic somite cell rearrangements lead to distinct waves of myotome growth. Development 2007, 134, 1253–1257. [Google Scholar] [CrossRef]
- Marty, I.; Robert, M.; Ronjat, M.; Bally, I.; Arlaud, G.; Villaz, M. Localization of the N-terminal and C-terminal ends of triadin with respect to the sarcoplasmic reticulum membrane of rabbit skeletal muscle. Biochem. J. 1995, 307 Pt 3, 769–774. [Google Scholar] [CrossRef]
- Marty, I.; Fauré, J.; Fourest-Lieuvin, A.; Vassilopoulos, S.; Oddoux, S.; Brocard, J. Triadin: What possible function 20 years later? J. Physiol. 2009, 587, 3117–3121. [Google Scholar] [CrossRef]
- Shen, X.; Franzini-Armstrong, C.; Lopez, J.R.; Jones, L.R.; Kobayashi, Y.M.; Wang, Y.; Kerrick, W.G.; Caswell, A.H.; Potter, J.D.; Miller, T.; et al. Triadins modulate intracellular Ca(2+) homeostasis but are not essential for excitation-contraction coupling in skeletal muscle. J. Biol. Chem. 2007, 282, 37864–37874. [Google Scholar] [CrossRef]
- Terentyev, D.; Cala, S.E.; Houle, T.D.; Viatchenko-Karpinski, S.; Gyorke, I.; Terentyeva, R.; Williams, S.C.; Gyorke, S. Triadin overexpression stimulates excitation-contraction coupling and increases predisposition to cellular arrhythmia in cardiac myocytes. Circ. Res. 2005, 96, 651–658. [Google Scholar] [CrossRef]
- Jin, D.; Ni, T.T.; Jia, H.; Rellinger, E.; Tao, P.Z. Promoter analysis of ventricular myosin heavy chain (vmhc) in zebrafish embryos. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009, 238, 1760–1767. [Google Scholar] [CrossRef]
- Zhang, R.; Han, P.; Yang, H.; Ouyang, K.; Lee, D.; Lin, Y.-F.; Ocorr, K.; Kang, G.; Chen, J.; Stainier, D.Y.R.; et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013, 498, 497–501. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L.; Gupta, V.A.; Beggs, A.H. Bridging integrator 1 (Bin1) deficiency in zebrafish results in centronuclear myopathy. Hum. Mol. Genet. 2014, 23, 3566–3578. [Google Scholar] [CrossRef]
- Kawahara, G.; Guyon, J.R.; Nakamura, Y.; Kunkel, L.M. Zebrafish models for human FKRP muscular dystrophies. Hum. Mol. Genet. 2010, 19, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L.; Beggs, A.H.; Gupta, V.A. Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J. Vis. Exp. JoVE 2013, e50925. [Google Scholar] [CrossRef]
- Yelon, D.; Horne, S.A.; Stainier, D.Y. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999, 214, 23–37. [Google Scholar] [CrossRef]
- Li, J.; Yue, Y.; Dong, X.; Jia, W.; Li, K.; Liang, D.; Dong, Z.; Wang, X.; Nan, X.; Zhang, Q.; et al. Zebrafish foxc1a plays a crucial role in early somitogenesis by restricting the expression of aldh1a2 directly. J. Biol. Chem. 2015, 290, 10216–10228. [Google Scholar] [CrossRef]
- Jowett, T.; Lettice, L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994, 10, 73–74. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B.; Schilling, T.F.; Postlethwait, J.H. Structure of the zebrafish snaill gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 1993, 119, 1203–1215. [Google Scholar] [CrossRef]
- Mastrodonato, V.; Beznoussenko, G.; Mironov, A.; Ferrari, L.; Deflorian, G.; Vaccari, T. A genetic model of CEDNIK syndrome in zebrafish highlights the role of the SNARE protein Snap29 in neuromotor and epidermal development. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Frise, V.; Longair, M.; Pietzsch, T.; Pietzsch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Gierten, J.; Pylatiuk, C.; Hammouda, O.T.; Schock, C.; Stegmaier, J.; Wittbrodt, J.; Gehrig, J.; Loosli, F. Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Kossack, M.; Hein, S.; Juergensen, L.; Siragusa, M.; Benz, A.; Katus, H.A.; Most, P.; Hassel, D. Induction of cardiac dysfunction in developing and adult zebrafish by chronic isoproterenol stimulation. J. Mol. Cell. Cardiol. 2017, 108, 95–105. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecchi, V.M.; Spreafico, M.; Brix, A.; Santoni, A.; Sala, S.; Pistocchi, A.; Marozzi, A.; Di Resta, C. Generation of a Triadin KnockOut Syndrome Zebrafish Model. Int. J. Mol. Sci. 2021, 22, 9720. https://doi.org/10.3390/ijms22189720
Vecchi VM, Spreafico M, Brix A, Santoni A, Sala S, Pistocchi A, Marozzi A, Di Resta C. Generation of a Triadin KnockOut Syndrome Zebrafish Model. International Journal of Molecular Sciences. 2021; 22(18):9720. https://doi.org/10.3390/ijms22189720
Chicago/Turabian StyleVecchi, Vanilla Martina, Marco Spreafico, Alessia Brix, Anna Santoni, Simone Sala, Anna Pistocchi, Anna Marozzi, and Chiara Di Resta. 2021. "Generation of a Triadin KnockOut Syndrome Zebrafish Model" International Journal of Molecular Sciences 22, no. 18: 9720. https://doi.org/10.3390/ijms22189720
APA StyleVecchi, V. M., Spreafico, M., Brix, A., Santoni, A., Sala, S., Pistocchi, A., Marozzi, A., & Di Resta, C. (2021). Generation of a Triadin KnockOut Syndrome Zebrafish Model. International Journal of Molecular Sciences, 22(18), 9720. https://doi.org/10.3390/ijms22189720






