Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future
Abstract
:1. Introduction
2. Diagnostics
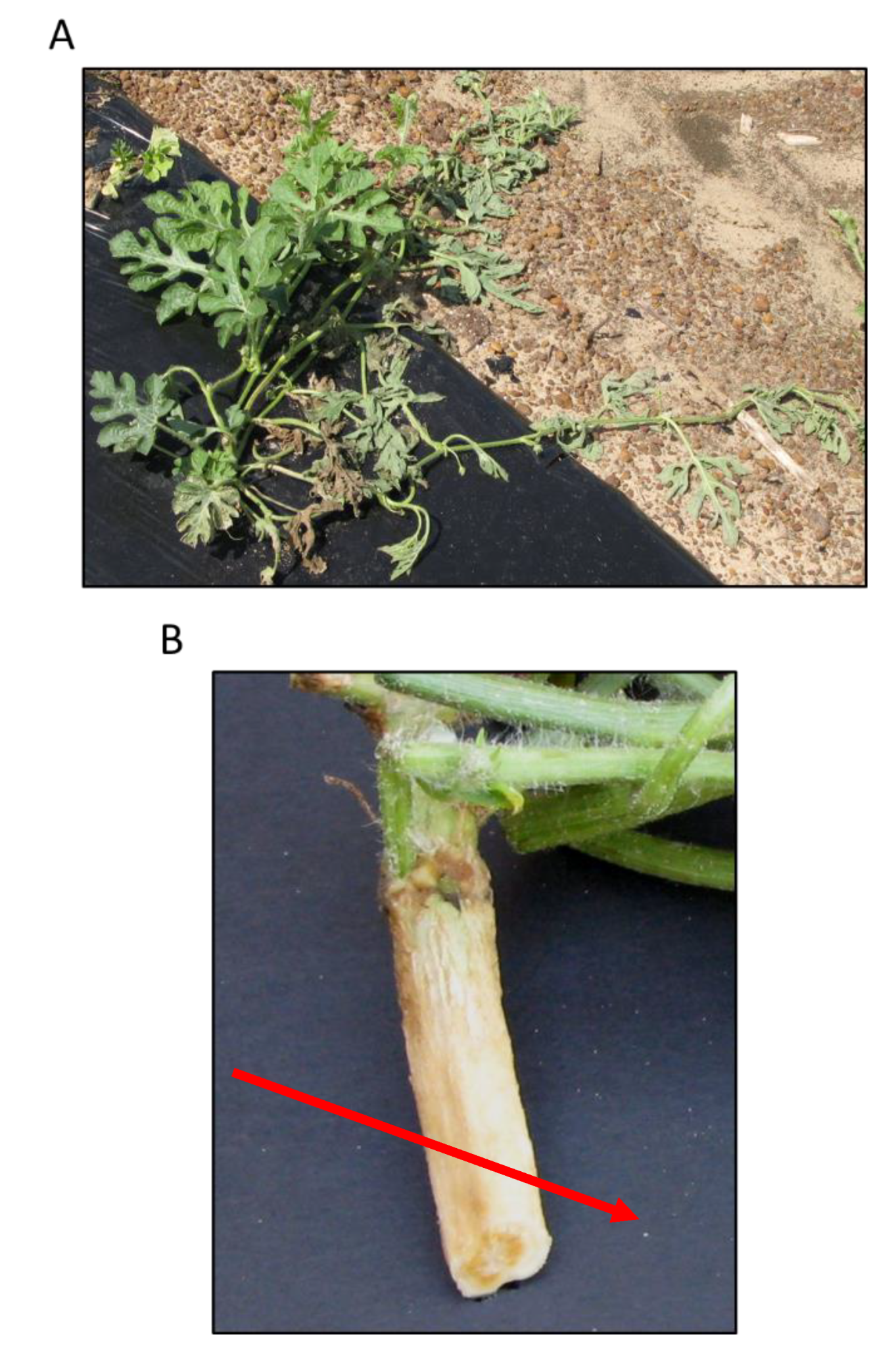
2.1. Bioassays
2.2. Molecular Assays
2.3. Evaluation of Diagnostic Methods
2.4. Genomics Diagnostics
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Fon | Fusarium oxysporum f. sp. niveum |
| SIX | Secreted in Xylem |
| FOSC | Fusarium oxysporum species complex |
| PCR | Polymerase chain reaction |
| RAPD | Random amplification of polymorphic DNA |
| PAMP | Pathogen associated molecular patterns |
| FOL | Fusarium oxysporum f. sp. lycopersici |
| BLAST | Basic local alignment search tool |
| MITE | Miniature Inverted-repeat Transposable Elements |
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef] [Green Version]
- Fravel, D.; Olivain, C.; Alabouvette, C. Fusarium oxysporum and its biocontrol. New Phytol. 2003, 157, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Perchepied, L.; Pitrat, M. Polygenic inheritance of partial resistance to Fusarium oxysporum f. sp. melonis race 1.2 in melon. Phytopathology 2004, 94, 1331–1336. [Google Scholar] [CrossRef] [Green Version]
- Lyons, R.; Stiller, J.; Powell, J.; Rusu, A.; Manners, J.M.; Kazan, K. Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS ONE 2015, 10, e0121902. [Google Scholar] [CrossRef] [PubMed]
- Gordon, T.; Martyn, R. The evolutionary biology of Fusarium oxysporum. Ann. Rev. Phytopathol. 1997, 35, 111–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recorbet, G.; Steinberg, C.; Olivain, C.; Edel, V.; Trouvelot, S.; Dumas-Gaudot, E.; Gianinazzi, S.; Alabouvette, C. Wanted: Pathogenesis-related marker molecules for Fusarium oxysporum. New Phytol. 2003, 159, 73–92. [Google Scholar] [CrossRef]
- Henry, P.M.; Pincot, D.D.; Jenner, B.N.; Borrero, C.; Aviles, M.; Nam, M.H.; Epstein, L.; Knapp, S.J.; Gordon, T.R. Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporum f. sp. fragariae. New Phytol. 2021, 230, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, P.; Fokkens, L.; Ayukawa, Y.; van der Gragt, M.; Ter Horst, A.; Brankovics, B.; Houterman, P.M.; Arie, T.; Rep, M. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 2017, 7, 9042. [Google Scholar] [CrossRef]
- Petkar, A.; Harris-Shultz, K.; Wang, H.; Brewer, M.T.; Sumabat, L.; Ji, P. Genetic and phenotypic diversity of Fusarium oxysporum f. sp. niveum populations from watermelon in the southeastern United States. PLoS ONE 2019, 14, e0219821. [Google Scholar] [CrossRef] [Green Version]
- Roberts, P.; Dufault, N.; Hochmuth, R.; Vallad, G.; Paret, M. [PP352] Fusarium Wilt (Fusarium oxysporum f. sp. niveum) of Watermelon. EDIS 2019, 2019, 4. [Google Scholar] [CrossRef]
- Ramos, B.; López, G.; Molina, A. Development of a Fusarium oxysporum f. sp. melonis functional GFP fluorescence tool to assist melon resistance breeding programmes. Plant Pathol. 2015, 64, 1349–1357. [Google Scholar] [CrossRef]
- Martyn, R.D. Fusarium wilt of watermelon: 120 years of research. Hortic. Rev. 2014, 42, 349–442. [Google Scholar] [CrossRef]
- Kleczewski, N.M.; Egel, D.S. A diagnostic guide for Fusarium wilt of watermelon. Plant Health Prog. 2011, 12, 27. [Google Scholar] [CrossRef] [Green Version]
- Larkin, R.; Hopkins, D.; Martin, F. Ecology of Fusarium oxysporum f. sp. niveum in soils suppressive and conducive to Fusarium wilt of watermelon. Phytopathology 1993, 83, 1105–1116. [Google Scholar]
- Peng, H.; Sivasithamparam, K.; Turner, D. Chlamydospore germination and Fusarium wilt of banana plantlets in suppressive and conducive soils are affected by physical and chemical factors. Soil Biol. Biochem. 1999, 31, 1363–1374. [Google Scholar] [CrossRef]
- Akhter, A.; Hage-Ahmed, K.; Soja, G.; Steinkellner, S. Potential of Fusarium wilt-inducing chlamydospores, in vitro behaviour in root exudates and physiology of tomato in biochar and compost amended soil. Plant Soil 2016, 406, 425–440. [Google Scholar] [CrossRef] [Green Version]
- El Mahjoub, M.; Le Picard, D.; Moreau, M. Origin of tyloses in melon (Cucumis melo L.) in response to a vascular fusarium. IAWA J. 1984, 5, 307–311. [Google Scholar] [CrossRef]
- VanderMolen, G.; Beckman, C.; Rodehorst, E. The ultrastructure of tylose formation in resistant banana following inoculation with Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 1987, 31, 185–200. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, J.; Liu, G.; Yao, X.; Li, P.; Yang, X. Characterization of the watermelon seedling infection process by Fusarium oxysporum f. sp. niveum. Plant Pathol. 2015, 64, 1076–1084. [Google Scholar] [CrossRef]
- Pariaud, B.; Ravigné, V.; Halkett, F.; Goyeau, H.; Carlier, J.; Lannou, C. Aggressiveness and its role in the adaptation of plant pathogens. Plant Pathol. 2009, 58, 409–424. [Google Scholar] [CrossRef]
- Zhou, X.; Everts, K.; Bruton, B. Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Dis. 2010, 94, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Fulton, J.C.; Amaradasa, B.S.; Ertek, T.S.; Iriarte, F.B.; Sanchez, T.; Ji, P.; Paret, M.L.; Hudson, O.; Ali, M.E.; Dufault, N.S. Phylogenetic and phenotypic characterization of Fusarium oxysporum f. sp. niveum isolates from Florida-grown watermelon. PLoS ONE 2021, 16, e0248364. [Google Scholar] [CrossRef]
- Hudson, O.; Waliullah, S.; Fulton, J.C.; Ji, P.; Dufault, N.S.; Keinath, A.; Ali, M.E. Marker Development for Differentiation of Fusarium oxysporum f. sp. niveum Race 3 from Races 1 and 2. Int. J. Mol. Sci. 2021, 22, 822. [Google Scholar] [CrossRef]
- Egel, D.; Martyn, R. Fusarium wilt of watermelon and other cucurbits. Plant Health Instr. 2007, 10, 1094. [Google Scholar]
- Keinath, A.P.; DuBose, V.B.; Katawczik, M.M.; Wechter, W.P. Identifying Races of Fusarium oxysporum f. sp. niveum in South Carolina Recovered From Watermelon Seedlings, Plants, and Field Soil. Plant Dis. 2020, 104, 2481–2488. [Google Scholar] [CrossRef] [PubMed]
- Gastner, M.T.; Seguy, V.; More, P. Fast flow-based algorithm for creating density-equalizing map projections. Proc. Natl. Acad. Sci. USA 2018, 115, E2156–E2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Everts, K. Characterization of a regional population of Fusarium oxysporum f. sp. niveum by race, cross pathogenicity, and vegetative compatibility. Phytopathology 2007, 97, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Everts, K. Races and inoculum density of Fusarium oxysporum f. sp. niveum in commercial watermelon fields in Maryland and Delaware. Plant Dis. 2003, 87, 692–698. [Google Scholar] [CrossRef] [Green Version]
- Kemble, J.M.; Meadows, I.; Jennings, K.; Walgenbach, J.; Wszelaki, A.L. (Eds.) Southeastern U.S. Vegetable Crop Handbook 2021, 22nd ed.; Great American Media Services: Sparta, MI, USA, 2021; p. 372. [Google Scholar]
- Coolong, B.D.T. Fusarium Wilt of Watermelon in Georgia; University of Georgia Extension: Athens, GA, USA, 2017; Available online: https://edis.ifas.ufl.edu/publication/PP352 (accessed on 24 October 2019).
- Zhang, Z.; Zhang, J.; Wang, Y.; Zheng, X. Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues and soil. FEMS Microbiol. Lett. 2005, 249, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-H.; Chen, K.-S.; Chang, J.-Y.; Wan, Y.-L.; Hsu, C.-C.; Huang, J.-W.; Chang, P.-F.L. Development of the molecular methods for rapid detection and differentiation of Fusarium oxysporum and F. oxysporum f. sp. niveum in Taiwan. New Biotechnol. 2010, 27, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Buller, S.; Inglis, D.; Miles, C. Plant growth, fruit yield and quality, and tolerance to verticillium wilt of grafted watermelon and tomato in field production in the Pacific Northwest. HortScience 2013, 48, 1003–1009. [Google Scholar] [CrossRef]
- Van Dam, P.; de Sain, M.; Ter Horst, A.; van der Gragt, M.; Rep, M. Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Appl. Environ. Microbiol. 2018, 84, e01868-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, X.; Zhao, X.; Ling, K.-S.; Levi, A.; Sun, Y.; Fan, M. The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum-watermelon pathosystem. Sci. Rep. 2016, 6, 28146. [Google Scholar] [CrossRef]
- Chang, W.; Li, H.; Chen, H.; Qiao, F.; Zeng, H. Identification of mimp-associated effector genes in Fusarium oxysporum f. sp. cubense race 1 and race 4 and virulence confirmation of a candidate effector gene. Microbiol. Res. 2020, 232, 126375. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S.; Di Pietro, A.; Daboussi, M.J.; Wahab, H.A.; Vasnier, C.; Roncero, M.I.G.; Dufresne, M.; Hera, C. Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large-scale transposon tagging. Mol. Plant Pathol. 2009, 10, 95–107. [Google Scholar] [CrossRef]
- Lévesque, C.A. Molecular methods for detection of plant pathogens—What is the future? Can. J. Plant Pathol. 2001, 23, 333–336. [Google Scholar] [CrossRef]
- Hadidi, A.; Levy, L.; Podleckis, E. Polymerase chain reaction technology in plant pathology. In Molecular Methods in Plant Pathology; CRC Press: Boca Raton, FL, USA, 2017; pp. 167–187. [Google Scholar] [CrossRef]
- Singh, U.S.; Singh, R.P. Molecular Methods in Plant Pathology; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar] [CrossRef]
- Keinath, A.; DuBose, V. First report of Fusarium oxysporum f. sp niveum race 2 in South Carolina watermelon fields. In Phytopathology; Amer Phytopathological Soc.: St Paul, MN, USA, 2009; Volume 99, p. S63. [Google Scholar]
- Poland, J.A.; Balint-Kurti, P.J.; Wisser, R.J.; Pratt, R.C.; Nelson, R.J. Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 2009, 14, 21–29. [Google Scholar] [CrossRef]
- Million, C.R.; Wijeratne, S.; Cassone, B.J.; Lee, S.; Rouf Mian, M.; McHale, L.K.; Dorrance, A.E. Hybrid genome assembly of a major quantitative disease resistance locus in soybean toward Fusarium graminearum. Plant Genome 2019, 12, 180102. [Google Scholar] [CrossRef]
- Quesada, T.; Gopal, V.; Cumbie, W.P.; Eckert, A.J.; Wegrzyn, J.L.; Neale, D.B.; Goldfarb, B.; Huber, D.A.; Casella, G.; Davis, J.M. Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.). Genetics 2010, 186, 677–686. [Google Scholar] [CrossRef] [Green Version]
- Bani, M.; Pérez-De-Luque, A.; Rubiales, D.; Rispail, N. Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp. pisi in pea. Front. Plant Sci. 2018, 9, 199. [Google Scholar] [CrossRef]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef]
- Houterman, P.M.; Speijer, D.; Dekker, H.L.; de Koster, C.G.; Cornelissen, B.J.; Rep, M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007, 8, 215–221. [Google Scholar] [CrossRef]
- Gawehns, F.; Houterman, P.; Ichou, F.A.; Michielse, C.; Hijdra, M.; Cornelissen, B.; Rep, M.; Takken, F. The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Sain, M.; Rep, M. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 2015, 16, 23970–23993. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Gardiner, D.M.; Kazan, K.; Manners, J.M. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant Microbe Interact. 2012, 25, 180–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalhais, L.C.; Henderson, J.; Rincon-Florez, V.A.; O’Dwyer, C.; Czislowski, E.; Aitken, E.A.; Drenth, A. Molecular diagnostics of banana Fusarium wilt targeting Secreted-in-Xylem genes. Front. Plant Sci. 2019, 10, 547. [Google Scholar] [CrossRef]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, A.; Rep, M.; Wang, B.; Ashton, A.; Dodds, P.; Ellis, J. Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum. Plant Pathol. 2011, 60, 232–243. [Google Scholar] [CrossRef]
- Van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef]
- Houterman, P.M.; Cornelissen, B.J.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, e1000061. [Google Scholar] [CrossRef] [Green Version]
- Houterman, P.M.; Ma, L.; Van Ooijen, G.; De Vroomen, M.J.; Cornelissen, B.J.; Takken, F.L.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’Neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.; Batley, J.; Aitken, E.A. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.M.; Houterman, P.M.; Schreiver, I.; Ma, L.; Amyotte, S.; Chellappan, B.; Boeren, S.; Takken, F.L.; Rep, M. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013, 14, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua-Van, A.; Pamphile, J.; Langin, T.; Daboussi, M.-J. Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. MGG 2001, 264, 724–731. [Google Scholar] [CrossRef]
- Van Dam, P.; Rep, M. The distribution of miniature impala elements and SIX genes in the Fusarium genus is suggestive of horizontal gene transfer. J. Mol. Evol. 2017, 85, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Batson, A.M.; Fokkens, L.; Rep, M.; du Toit, L.J. Putative effector genes distinguish two pathogenicity groups of Fusarium oxysporum f. sp. spinaciae. Mol. Plant Microbe Interact. 2021, 34, 141–156. [Google Scholar] [CrossRef]
- Fulton, J.; Brawner, J.; Huguet-Tapia, J.; Smith, K.E.; Fernandez, R.; Dufault, N.S. Six de novo assemblies from pathogenic and non-pathogenic strains of Fusarium oxysporum f. sp. niveum. PhytoFrontiers 2021. [Google Scholar] [CrossRef]
- Hudson, O.; Hudson, D.; Ji, P.; Ali, M.E. Draft genome sequences of three Fusarium oxysporum f. sp. niveum isolates used in designing markers for race differentiation. Microbiol. Resour. Announc. 2020, 9, e01004-20. [Google Scholar] [CrossRef] [PubMed]




| Race 0 | Race 1 | Race 2 | Race 3 | Reference | |
|---|---|---|---|---|---|
| Sugar Baby | S | S | S | S | [1,3,4] |
| Black Diamond | S | S | S | S | [1,3,5] |
| Charleston Gray | R | S | S | S | [1,5] |
| Crimson Sweet | R | S | S | S | [1,5] |
| Mickey Lee | R | R | S | S | [4] |
| Dixielee * | R | S | S | S | [2] |
| Dixielee * | R | R | S | S | [3] |
| Allsweet * | R | S | S | S | [2] |
| Allsweet * | R | R | S | S | [1,3,5] |
| Calhoun Gray | R | R | S | S | [1,3,5] |
| PI-296341-FR | R | R | R | S | [1] |
| Purpose of Assay | Primers | Sequence (5′-3′) | Product Size (bp) | Source |
|---|---|---|---|---|
| Fon-specific primer | Fon-1 | CGATTAGCGAAGACATTCACAAGACT | 174 | Lin et al., 2010 [33] |
| Fon-2 | ACGGTCAAGAAGATGCAGGGTAAAGGT | |||
| Race 2 differentiating primer | FONSIX6F | CGCTCTTATCGCATCAATCT | 453 | Niu et al., 2016 [36] |
| FONSIX6R | GGGTTGACTGAGGTCGTGGT | |||
| Race 3 differentiating primer | FNR3F | CGGCTTTCCTCTGTCAGATAGT | 511 | Hudson et al., 2021 [24] |
| FNR3R | TAGTGAGGTCCATGCCACGAA |
| Diagnostic Technique | Advantages | Disadvantages |
|---|---|---|
| Field observation | Fastest, low training needs | Not accurate, no distinct race determination |
| Culturing and microscopy | Only basic lab instruments required. Fusarium sp. confirmation rapidly. | Skills for culturing, isolation, and microscopy required. Knowledge of morphology required. No species or race level determination possible |
| Greenhouse bioassay | Race identification possible. Highly controlled environment allowing for statistical analysis. Current standard for evaluation of Fon isolates | Culturing of Fon required. Specific cultivar’s seeds, controlled greenhouse environment, sterile locations required. Multiple replications needed for confirmation. Several months for final results |
| Molecular (PCR based) | Ability to rapidly determine species and often race level of Fon. Very fast to determine Fon races. | Access to molecular lab and molecular lab equipment. Training in all PCR methods required. Not confirmed to be able to distinguish all races. Unable to determine race if multiple genes involved. |
| Gene profile | Able to determine all possible variations of effectors present and active in a Fon isolate. Theoretically exact race determination possible even if resistance is quantitative. Once determined, the ability can be converted to conventional molecular methods (PCR). | Multiple gene sequencing of individual isolates required. Higher level of knowledge for sequencing analysis and multigene sequencing required. Instruments additionally more complex and expensive. More annotations and whole genome sequencing required. |
| Putative Race | 0 | 1 | 2 | 3 | Unknown | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate Name | 110407_3_1_1 | 150523 | R1 | R2 | 150524 | R3 | Fon002 | Fon005 | Fon010 | Fon013 | Fon015 | Fon019 | Fon020 | Fon021 | Fon037 |
| Accession number | GCA_01959 3455.1 [65] | GCA_01959 3445.1 [65] | GCA_0146 02815.1 [66] | GCA_0146 02775.1 [66] | GCA_0195 93505.1 [65] | GCA_0146 02795.1 [66] | GCA_0017 02745.1 [9] | GCA_0017 02505.1 [9] | GCA_0017 02785.1 [9] | GCA_0017 02775.1 [9] | GCA_0017 02795.1 [9] | GCA_0017 02715.1 [9] | GCA_0017 02805.1 [9] | GCA_0017 02865.1 [9] | GCA_0017 02845.1 [9] |
| SIX1 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX2 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX3 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX4 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX5 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX6 | Absent | Present | Present | Absent | Absent | Present | Absent | Present | Absent | Present | Absent | Absent | Present | Absent | Absent |
| SIX7 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX8 | Absent | Present | Present | Present | Present | Present | Absent | Present | Present | Present | Absent | Absent | Present | Present | Absent |
| SIX9 | Present | Present | Present | Present | Present | Present | Absent | Present | Present | Present | Present | Present | Present | Present | Absent |
| SIX10 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX11 | Absent | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present |
| SIX12 | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| SIX13 | Absent | Present | Present | Absent | Absent | Present | Present | Present | Present | Present | Present | Present | Present | Present | Present |
| SIX14 | Absent | Absent | Absent | Absent | Absent | Absent | Present | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudson, O.; Fulton, J.C.; Dong, A.K.; Dufault, N.S.; Ali, M.E. Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future. Int. J. Mol. Sci. 2021, 22, 9735. https://doi.org/10.3390/ijms22189735
Hudson O, Fulton JC, Dong AK, Dufault NS, Ali ME. Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future. International Journal of Molecular Sciences. 2021; 22(18):9735. https://doi.org/10.3390/ijms22189735
Chicago/Turabian StyleHudson, Owen, James C. Fulton, Alexi K. Dong, Nicholas S. Dufault, and Md Emran Ali. 2021. "Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future" International Journal of Molecular Sciences 22, no. 18: 9735. https://doi.org/10.3390/ijms22189735
APA StyleHudson, O., Fulton, J. C., Dong, A. K., Dufault, N. S., & Ali, M. E. (2021). Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future. International Journal of Molecular Sciences, 22(18), 9735. https://doi.org/10.3390/ijms22189735







