Ablation of KDM2A Inhibits Preadipocyte Proliferation and Promotes Adipogenic Differentiation
Abstract
:1. Introduction
2. Results
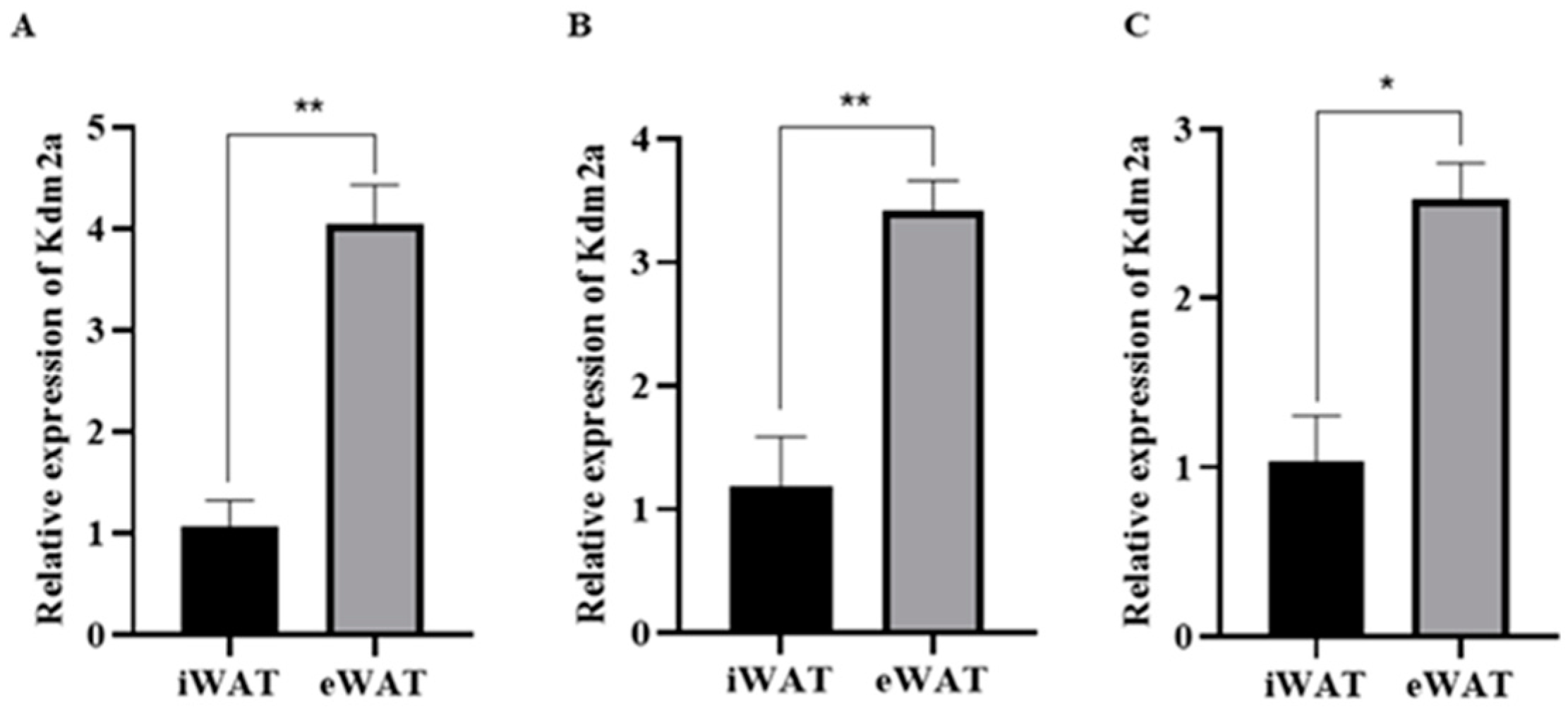
2.1. Expression Pattern of Histone Demethylase 2A (KDM2A) in White Adipose Tissues at Different Stages
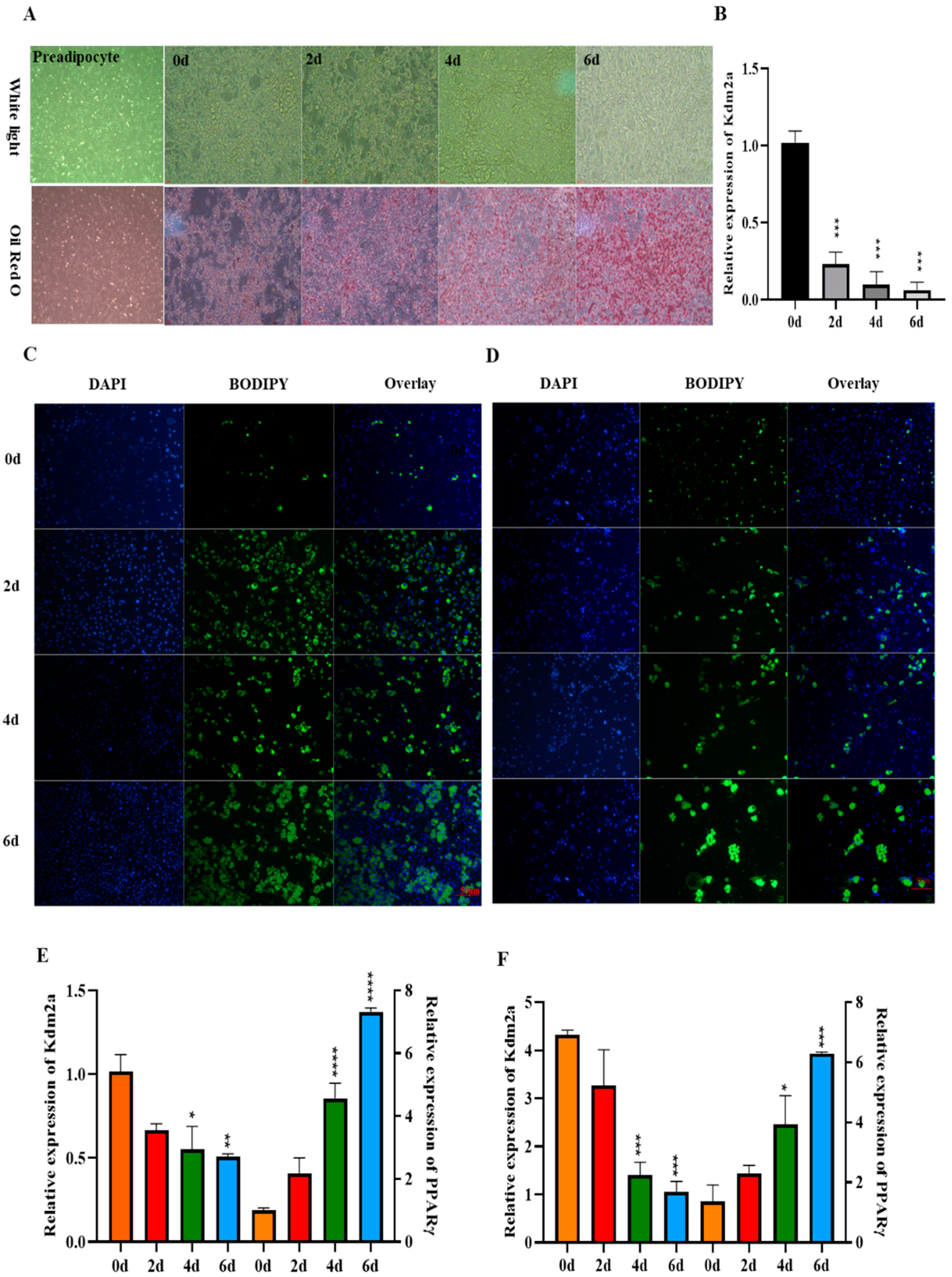
2.2. Expression Pattern of KDM2A during Adipogenic Differentiation
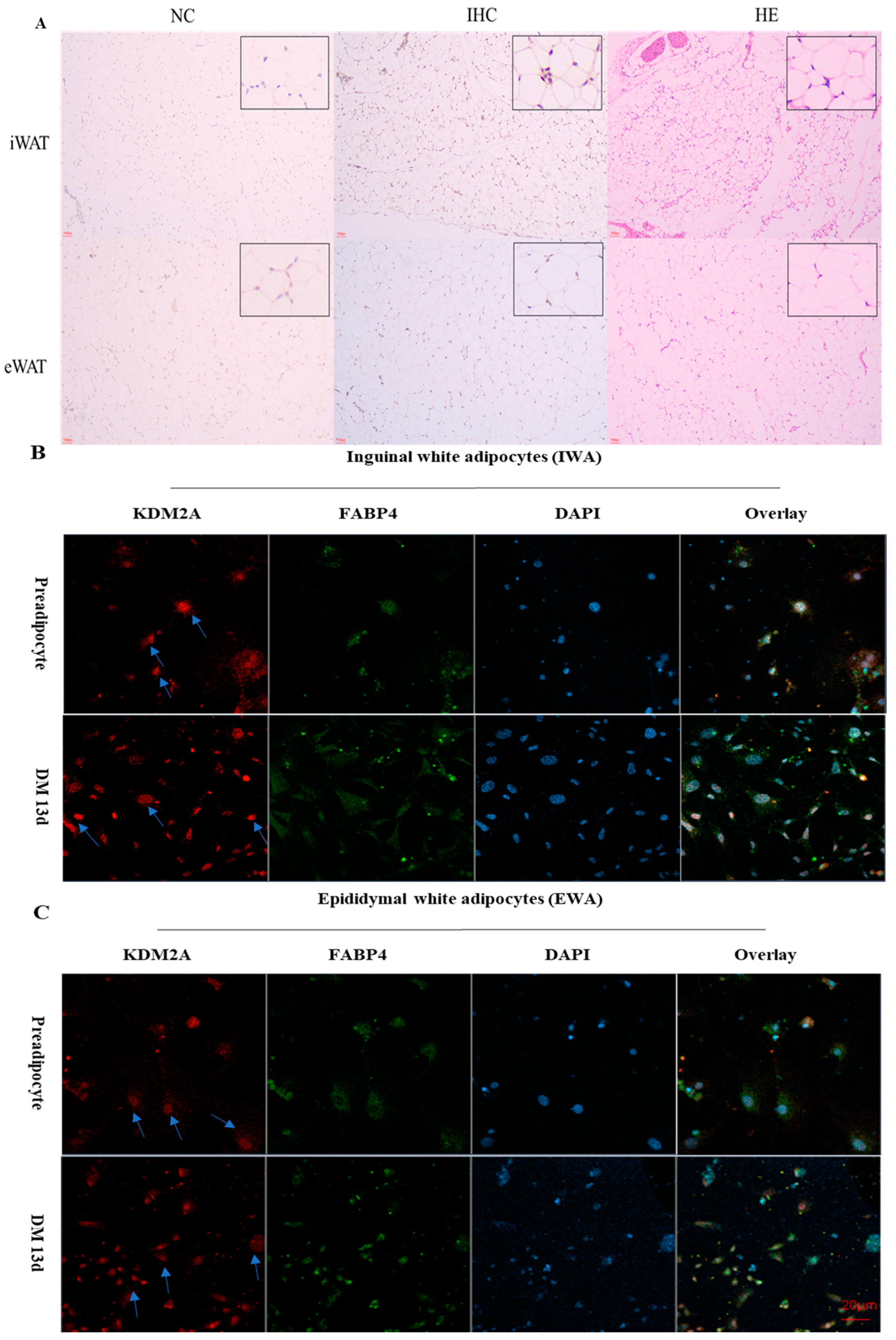
2.3. The Subcellular Location of KDM2A In Vivo and In Vitro
2.4. Knockdown of KDM2A Inhibits the Proliferation of 3T3-L1 Preadipocytes
2.5. Knockdown of KDM2A Promotes 3T3-L1 Preadipocyte Differentiation
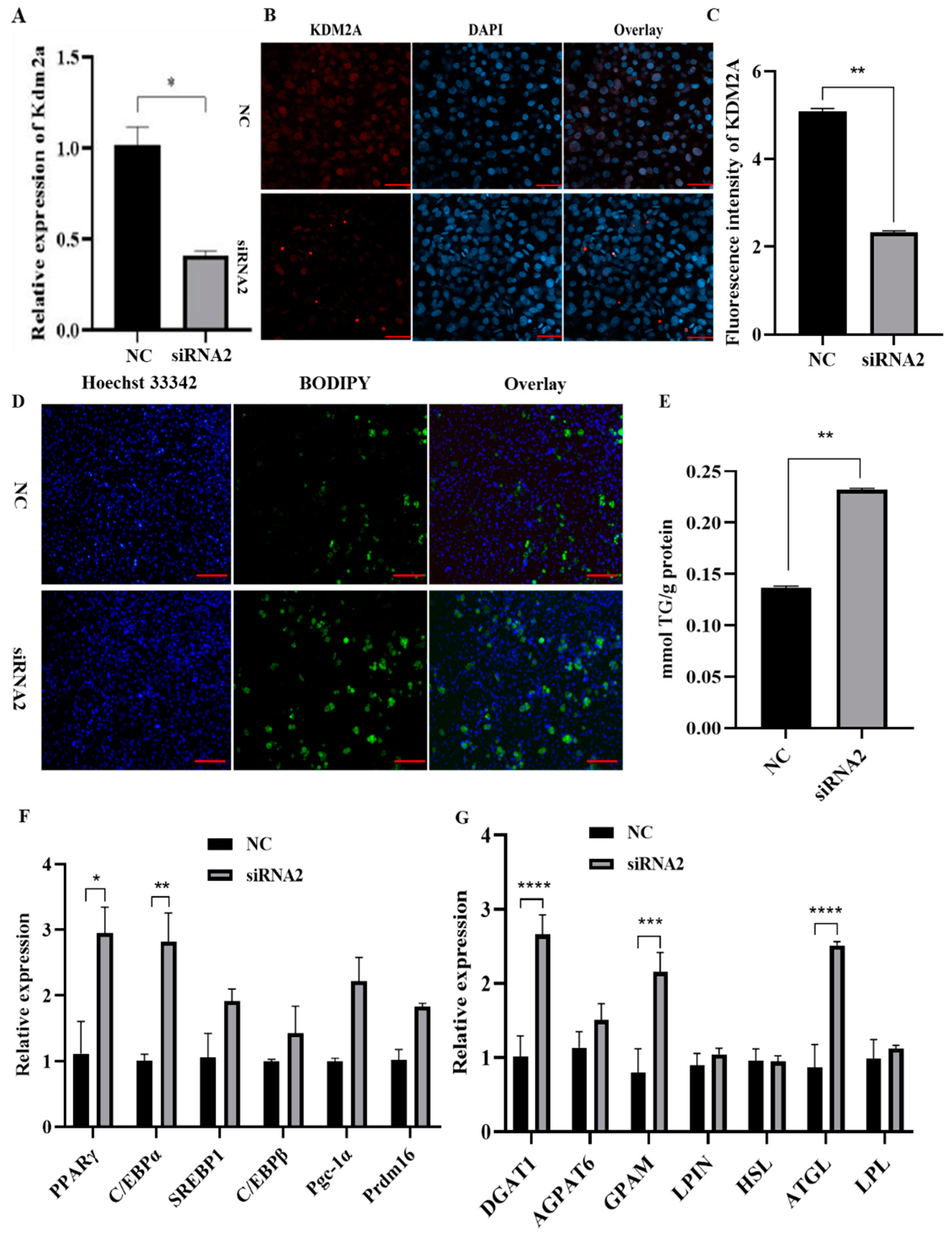
2.6. Deletion of KDM2A Promotes Primary Adipocytes Differentiation
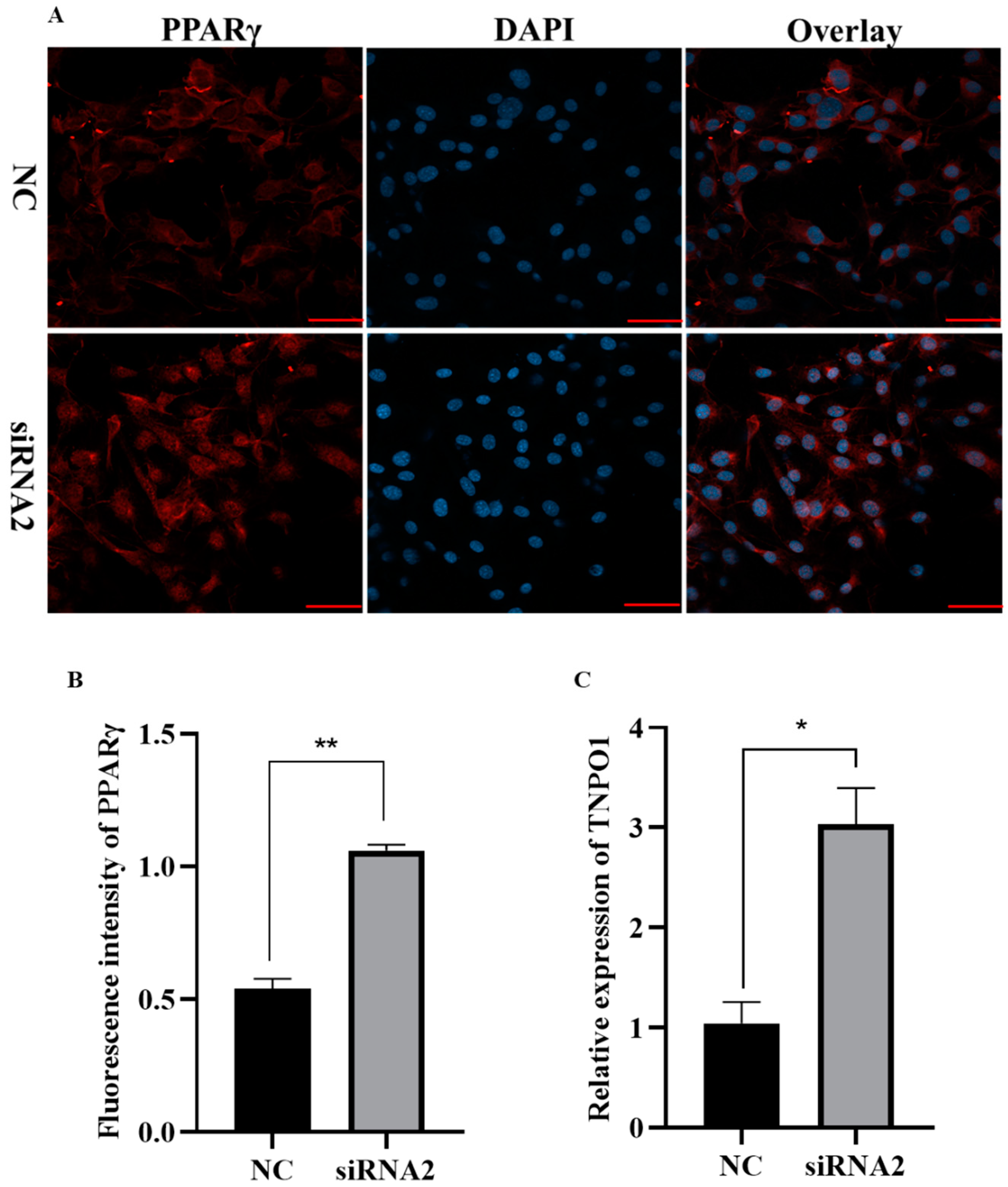
2.7. Knockdown of KDM2A Promotes Adipogenesis by Affecting Expression and Nuclear Translocation of PPARγ
3. Discussion
4. Materials and Methods
4.1. Animals and Cell Line
4.2. Primary Adipocyte Isolation, Culture and Differentiation
4.3. BODIPY and Oil Red O Staining
4.4. Immunofluorescence Staining
4.5. Total RNA Extraction and Quantitative Real-Time PCR (qPCR)
4.6. Chemical Synthesis of siRNA
4.7. Cell Transfection
4.8. EdU Staining
4.9. CCK-8 Detection
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Mariosa, D.; Carreras-Torres, R.; Martin, R.M.; Johansson, M.; Brennan, P. Commentary: What can Mendelian randomization tell us about causes of cancer? Int. J. Epidemiol. 2019, 48, 816–821. [Google Scholar] [CrossRef]
- Bastien, M.; Poirier, P.; Lemieux, I.; Despres, J.P. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog. Cardiovasc. Dis. 2014, 56, 369–381. [Google Scholar] [CrossRef]
- Gill, T.; King, L.; Caterson, I. Obesity prevention: Necessary and possible. A structured approach for effective planning. Proc. Nutr. Soc. 2005, 64, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Karpe, F.; Tan, G.D. Adipose tissue function in the insulin-resistance syndrome. Biochem. Soc. Trans. 2005, 33, 1045–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, J.; Goedecke, J.H.; Soderstrom, I.; Buren, J.; Alvehus, M.; Blomquist, C.; Jonsson, F.; Hayes, P.M.; Adams, K.; Dave, J.A.; et al. Depot- and ethnic-specific differences in the relationship between adipose tissue inflammation and insulin sensitivity. Clin. Endocrinol. 2011, 74, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Moustaid-Moussa, N. Secretory, endocrine and autocrine/paracrine function of the adipocyte. J. Nutr. 2000, 130, 3110S–3115S. [Google Scholar] [CrossRef]
- Smith, J.M.; Maas, J.A.; Garnsworthy, P.C.; Owen, M.R.; Coombes, S.; Pillay, T.S.; Barrett, D.A.; Symonds, M.E. Mathematical modeling of glucose homeostasis and its relationship with energy balance and body fat. Obesity 2009, 17, 632–639. [Google Scholar] [CrossRef]
- Lv, X.; Qiu, J.; Hao, T.; Zhang, H.; Jiang, H.; Tan, Y. HDAC inhibitor Trichostatin A suppresses adipogenesis in 3T3-L1 preadipocytes. Aging (Albany N. Y.) 2021, 13, 17489–17498. [Google Scholar] [CrossRef]
- Castellano-Castillo, D.; Ramos-Molina, B.; Cardona, F.; Queipo-Ortuno, M.I. Epigenetic regulation of white adipose tissue in the onset of obesity and metabolic diseases. Obes. Rev. 2020, 21, e13054. [Google Scholar] [CrossRef]
- Remely, M.; de la Garza, A.L.; Magnet, U.; Aumueller, E.; Haslberger, A.G. Obesity: Epigenetic regulation—Recent observations. Biomol. Concepts 2015, 6, 163–175. [Google Scholar] [CrossRef]
- Cui, L.; Fan, Q.; Cui, L.; Miao, J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol. 2008, 38, 1083–1097. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Okamoto, K.; Teye, K.; Umata, T.; Yamagiwa, N.; Suto, Y.; Zhang, Y.; Tsuneoka, M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010, 29, 1510–1522. [Google Scholar] [CrossRef] [Green Version]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Chen, C.L.; Huang, S.C.; Antonescu, C.R. Expanding the molecular signature of ossifying fibromyxoid tumors with two novel gene fusions: CREBBP-BCORL1 and KDM2A-WWTR1. Genes Chromosomes Cancer 2017, 56, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Wagner, K.W.; Alam, H.; Dhar, S.S.; Giri, U.; Li, N.; Wei, Y.K.; Cascone, T.; Giri, D.; Kim, J.H.; Ye, Y.Q.; et al. The histone demethylase KDM2A is a new promoter of tumorigenesis, drug target and negative prognostic biomarker for non-small cell lung cancer. Cancer Res. 2014, 74, 5146. [Google Scholar]
- Zhao, Y.L.; Chen, X.B.; Jiang, J.; Wan, X.C.; Wang, Y.F.; Xu, P. Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165856. [Google Scholar] [CrossRef]
- Huang, X.Y. Establishment of Specific KDM2A Knockout Mouse Model and Its Effect on Male Reproduction. Master’s Thesis, Southwest University for Nationalities, Chengdu, China, 2020. [Google Scholar]
- Boniakowski, A.; Kimball, A.; Davis, F.; Joshi, A.; Schaller, M.; den Dekker, A.; Kunkel, S.; Gallagher, K. Fatty Acid Binding Protein 4, FABP4, Causes Impaired Wound Healing in Diabetes. Arterioscler. Thromb. Vasc. Biol. 2018, 38, A255. [Google Scholar] [CrossRef]
- Yonekura, S.; Hirota, S.; Miyazaki, H.; Tokutake, Y. Subcellular Localization and Polymorphism of Bovine FABP4 in Bovine Intramuscular Adipocytes. Anim. Biotechnol. 2016, 27, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Ayers, S.D.; Nedrow, K.L.; Gillilan, R.E.; Noy, N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry 2007, 46, 6744–6752. [Google Scholar] [CrossRef]
- Lee, K.Y.; Russell, S.J.; Ussar, S.; Boucher, J.; Vernochet, C.; Mori, M.A.; Smyth, G.; Rourk, M.; Cederquist, C.; Rosen, E.D.; et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 2013, 62, 864–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.M.; Patel, T.P.; Altman, E.K.; Tugarinov, N.; Trivellin, G.; Yanovski, J.A. Characterization of the adiponectin promoter + Cre recombinase insertion in the Tg(Adipoq-cre)1Evdr mouse by targeted locus amplification and droplet digital PCR. Adipocyte 2021, 10, 21–27. [Google Scholar] [CrossRef]
- Chen, L.M.; Zhang, J.; Zou, Y.; Wang, F.X.; Li, J.Y.; Sun, F.; Luo, X.; Zhang, M.; Guo, Y.C.; Yu, Q.L.; et al. Kdm2a deficiency in macrophages enhances thermogenesis to protect mice against HFD-induced obesity by enhancing H3K36me2 at the Pparg locus. Cell Death Differ. 2021, 28, 1880–1899. [Google Scholar] [CrossRef] [PubMed]
- Teratani, T.; Tomita, K.; Toma-Fukai, S.; Nakamura, Y.; Itoh, T.; Shimizu, H.; Shiraishi, Y.; Sugihara, N.; Higashiyama, M.; Shimizu, T.; et al. Redox-dependent PPAR gamma/Tnpo1 complex formation enhances PPAR gamma nuclear localization and signaling. Free Radic. Biol. Med. 2020, 156, 45–56. [Google Scholar] [CrossRef]
- Noberini, R.; Restellini, C.; Savoia, E.O.; Bonaldi, T. Enrichment of histones from patient samples for mass spectrometry-based analysis of post-translational modifications. Methods 2020, 184, 19–28. [Google Scholar] [CrossRef]
- Zhang, X.D.; Saarinen, A.M.; Campbell, L.E.; De Filippis, E.A.; Liu, J. Regulation of Lipolytic Response and Energy Balance by Melanocortin 2 Receptor Accessory Protein (MRAP) in Adipocytes. Diabetes 2018, 67, 222–234. [Google Scholar] [CrossRef]
- Samimi, H.; Haghpanah, V.; Irani, S.; Arefian, E.; Sohi, A.N.; Fallah, P.; Soleimani, M. Transcript-level regulation of MALAT1-mediated cell cycle and apoptosis genes using dual MEK/Aurora kinase inhibitor “BI-847325” on anaplastic thyroid carcinoma. Daru-J. Pharm. Sci. 2019, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gross, N.; Li, Z.F.; Yin, G.F.; Zhong, Q.; Liu, C.; Huang, Z.G. Upregulation of BTF3 affects the proliferation, apoptosis, and cell cycle regulation in hypopharyngeal squamous cell carcinoma. Biomed. Pharmacother. 2019, 118, 109211. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.H.; Liang, D.Y.; Wang, Q.; Shen, J.H.; Liu, Q.H.; Peng, Y.B. Knockdown of KDM2A inhibits proliferation associated with TGF-beta expression in HEK293T cell. Mol. Cell. Biochem. 2019, 456, 95–104. [Google Scholar] [CrossRef]
- Wang, F.; Liang, S.B.; Liu, X.W.; Han, L.; Wang, J.Y.; Du, Q. LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR-342-3p in gastric cancer. Oncotargets Ther. 2018, 11, 6383–6394. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.L.; Li, J.Q.; Wang, Z.J.; Meng, J.; Wang, A.H.; Zhao, X.F.; Xu, Q.L.; Cai, Z.; Hu, Z.B. KDM2A Targets PFKFB3 for Ubiquitylation to Inhibit the Proliferation and Angiogenesis of Multiple Myeloma Cells. Front. Oncol. 2021, 11, 653788. [Google Scholar] [CrossRef] [PubMed]
- Burrell, J.A.; Boudreau, A.; Stephens, J.M. Latest advances in STAT signaling and function in adipocytes. Clin. Sci. 2020, 134, 629–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.H.; Sun, R.H.; Liu, L.P. Potentially critical roles of TNPO1, RAP1B, ZDHHC17, and PPM1B in the progression of coronary atherosclerosis through microarray data analysis. J. Cell. Biochem. 2019, 120, 4301–4311. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.l.; Xiong, Y.; Zhang, J.; Guo, Y.; Qin, W.C.; Xiong, X.R.; Zi, X.D.; Yin, S.; Li, J. Screening of reference genes for quantitative PCR in adipose deposition. Chin. J. Vet. Sci. 2020, 40, 2398–2404. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Yue, Y.; Zhao, D.; Ma, Y.; Xiong, Y.; Xiong, X.; Li, J. Ablation of KDM2A Inhibits Preadipocyte Proliferation and Promotes Adipogenic Differentiation. Int. J. Mol. Sci. 2021, 22, 9759. https://doi.org/10.3390/ijms22189759
Hua Y, Yue Y, Zhao D, Ma Y, Xiong Y, Xiong X, Li J. Ablation of KDM2A Inhibits Preadipocyte Proliferation and Promotes Adipogenic Differentiation. International Journal of Molecular Sciences. 2021; 22(18):9759. https://doi.org/10.3390/ijms22189759
Chicago/Turabian StyleHua, Yonglin, Yongqi Yue, Dan Zhao, Yan Ma, Yan Xiong, Xianrong Xiong, and Jian Li. 2021. "Ablation of KDM2A Inhibits Preadipocyte Proliferation and Promotes Adipogenic Differentiation" International Journal of Molecular Sciences 22, no. 18: 9759. https://doi.org/10.3390/ijms22189759
APA StyleHua, Y., Yue, Y., Zhao, D., Ma, Y., Xiong, Y., Xiong, X., & Li, J. (2021). Ablation of KDM2A Inhibits Preadipocyte Proliferation and Promotes Adipogenic Differentiation. International Journal of Molecular Sciences, 22(18), 9759. https://doi.org/10.3390/ijms22189759






