Identification of miR-199a-5p, miR-214-3p and miR-99b-5p as Fibrosis-Specific Extracellular Biomarkers and Promoters of HSC Activation
Abstract
:1. Introduction
2. Results
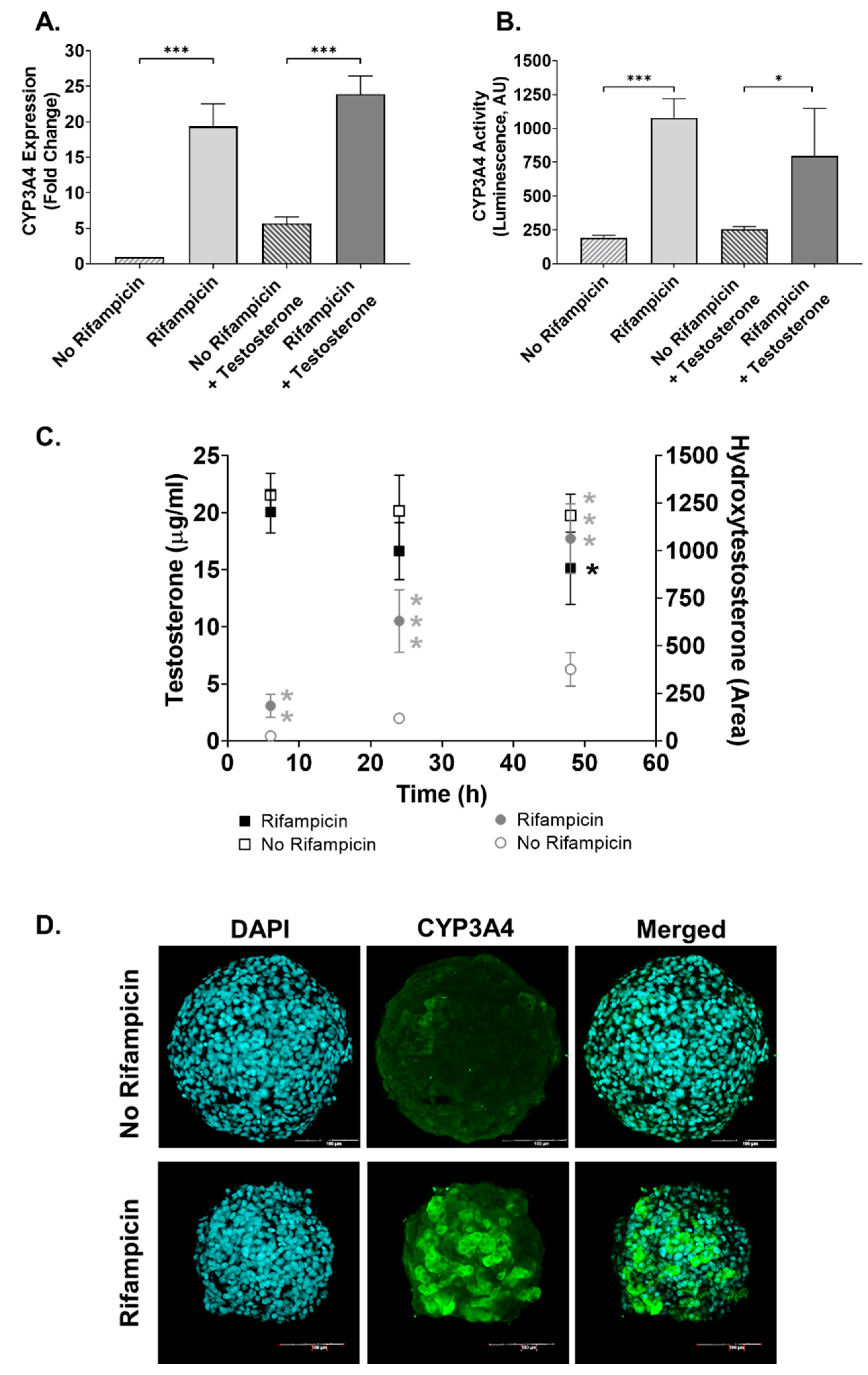
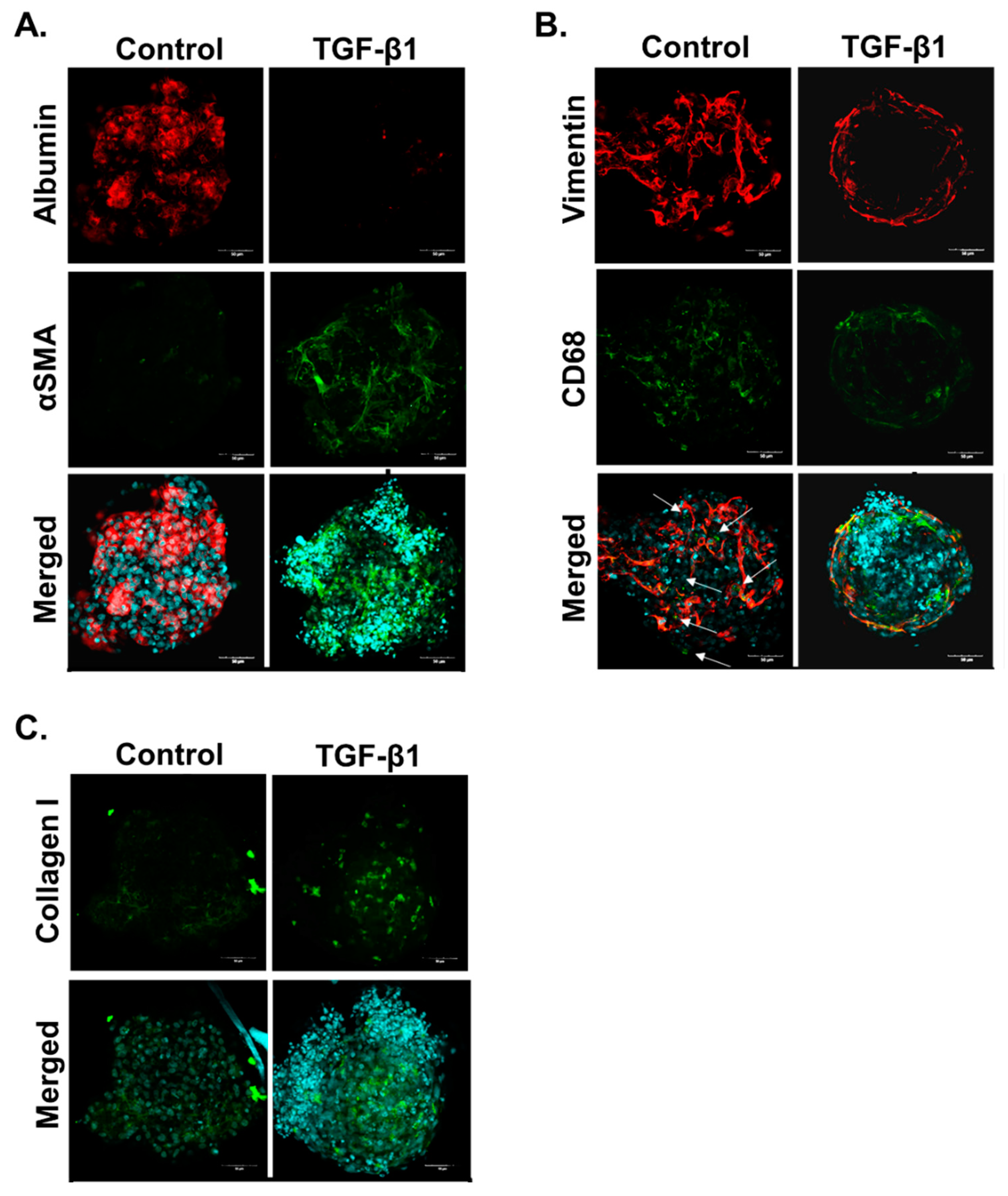
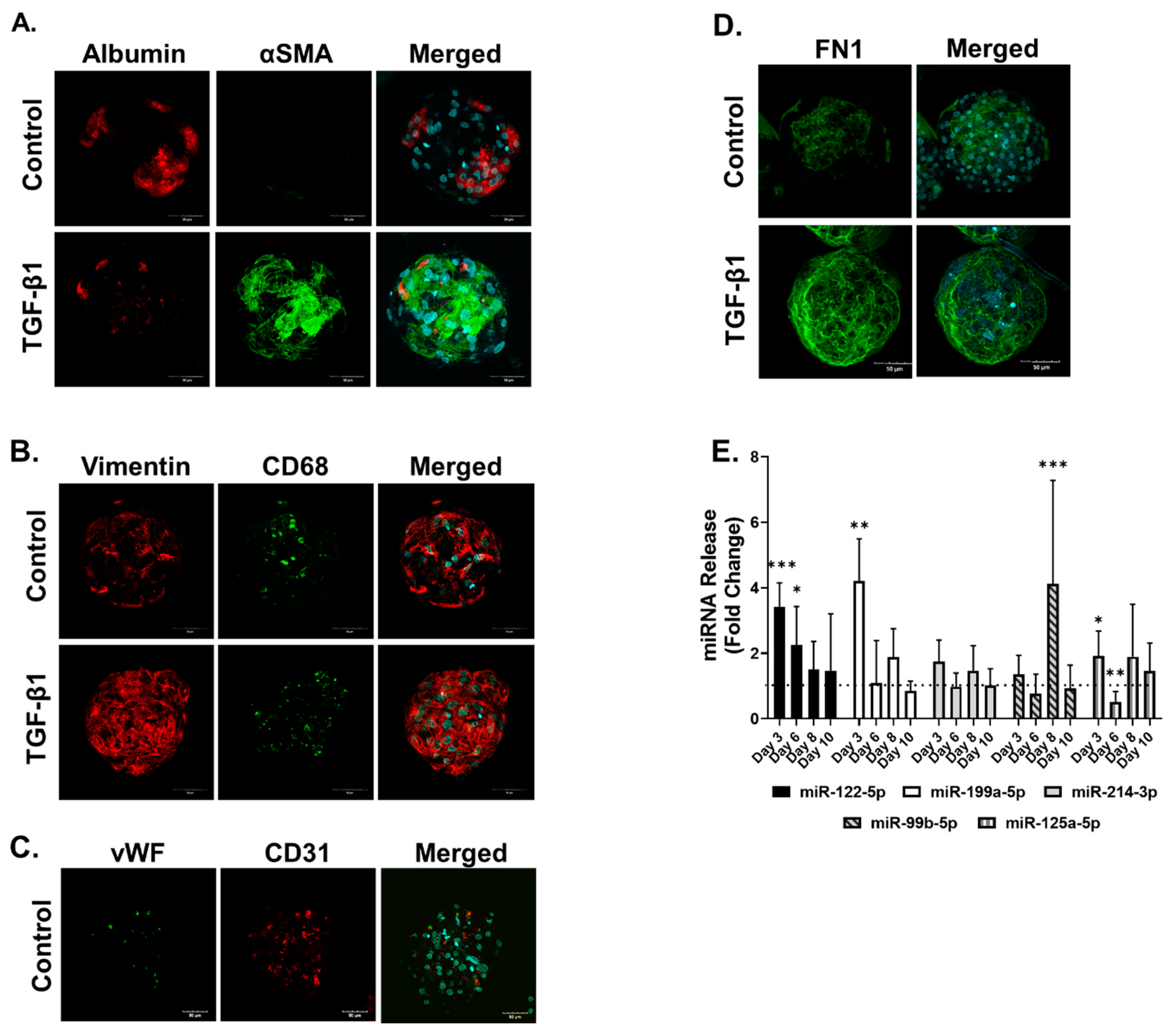
2.1. The 3D In Vitro Liver Model Maintains Metabolic Activity and Can Develop a Fibrotic Phenotype
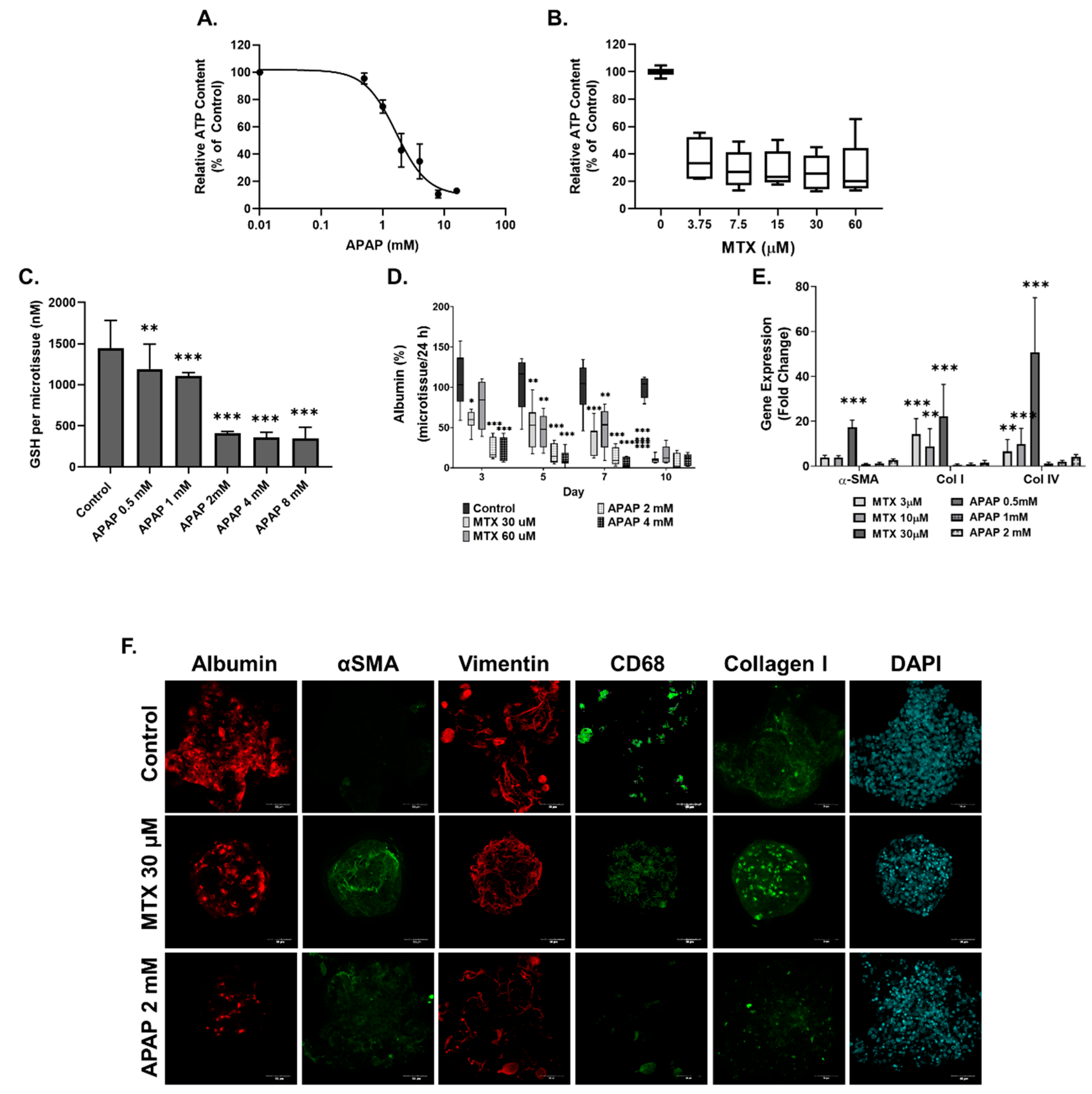
2.2. Methotrexate but Not Acetaminophen Induces Fibrosis in the MTs
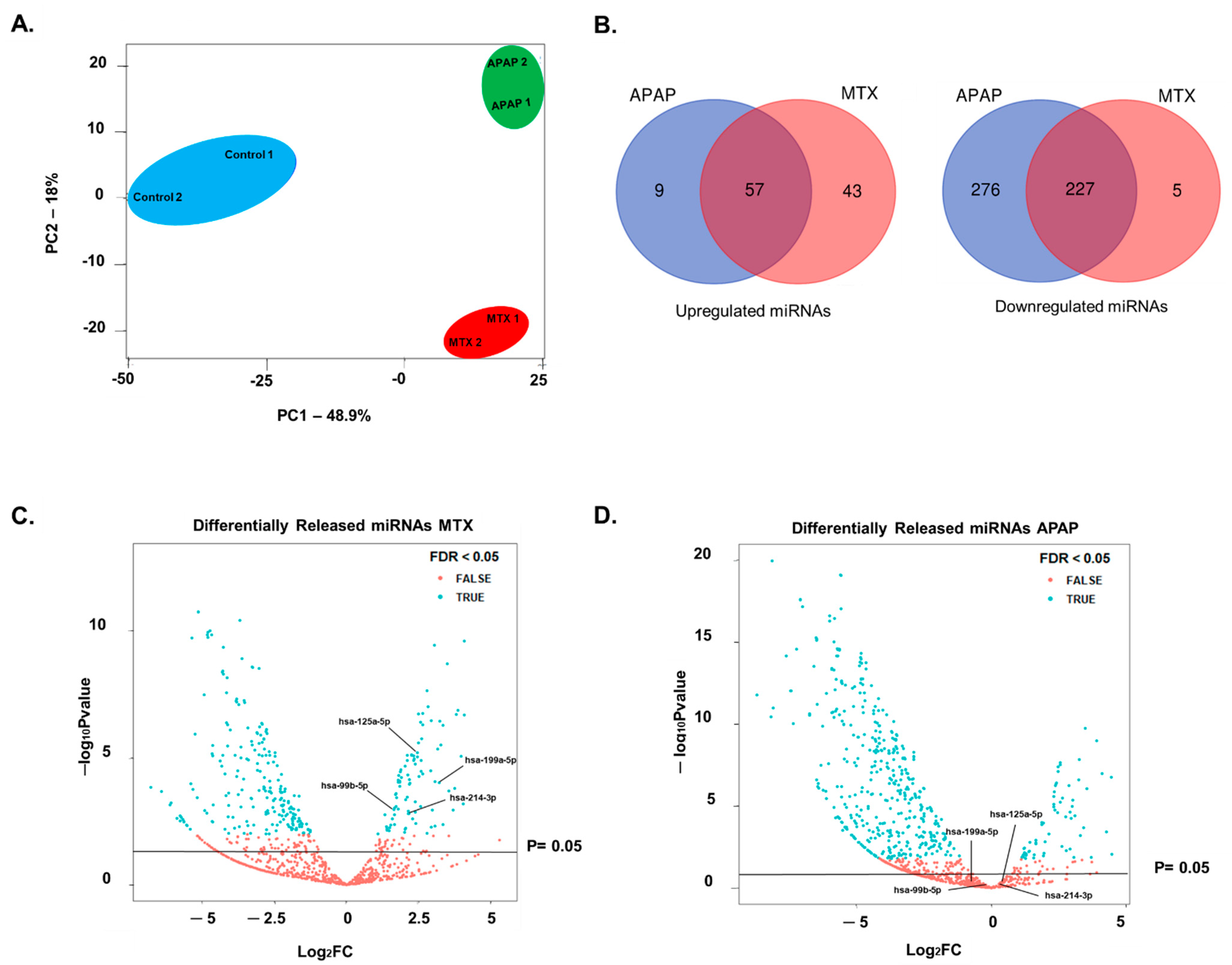
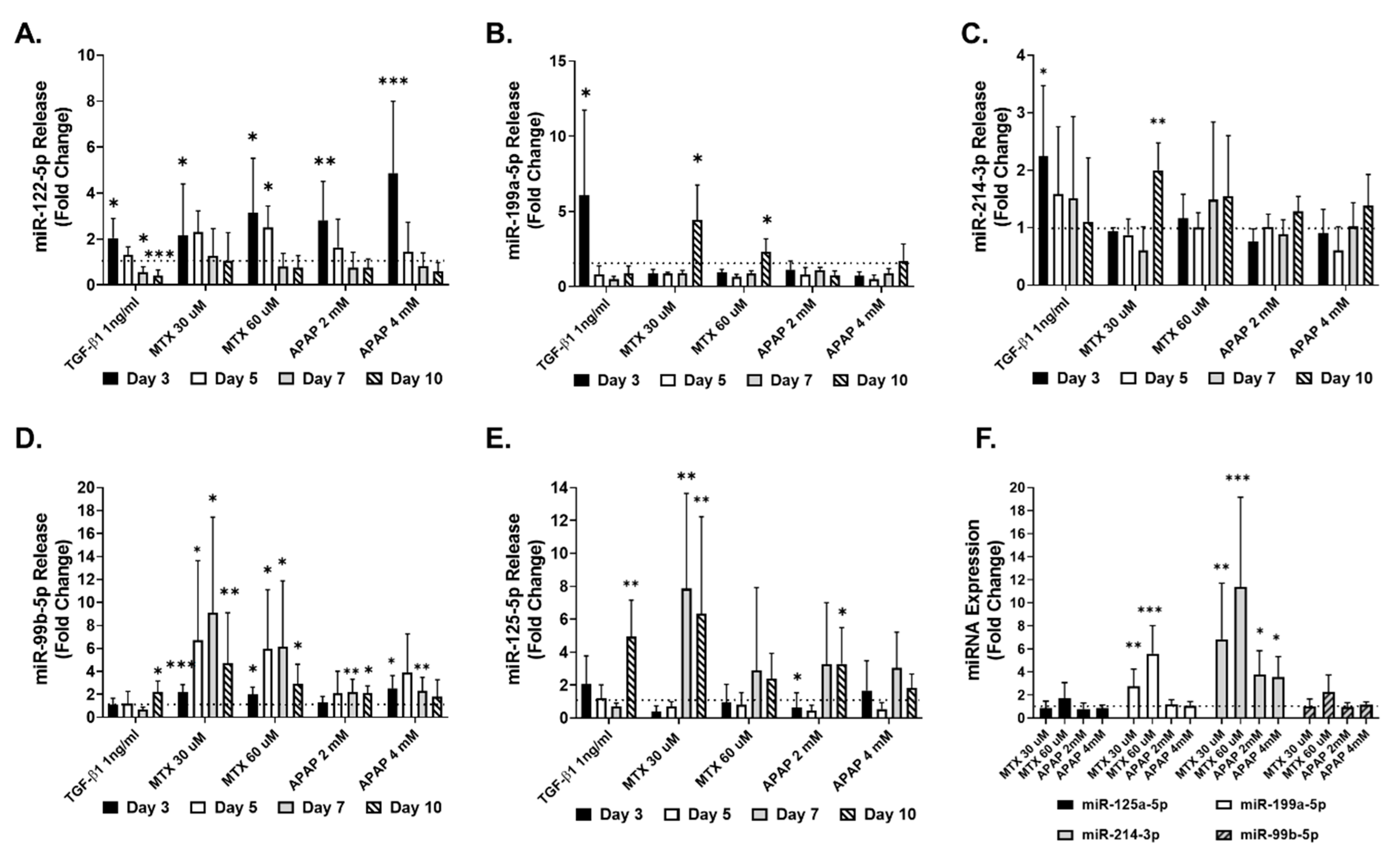
2.3. Identification of Fibrosis-Specific miRNAs
2.4. Translation from Human Cell Lines to Human Primary Cells
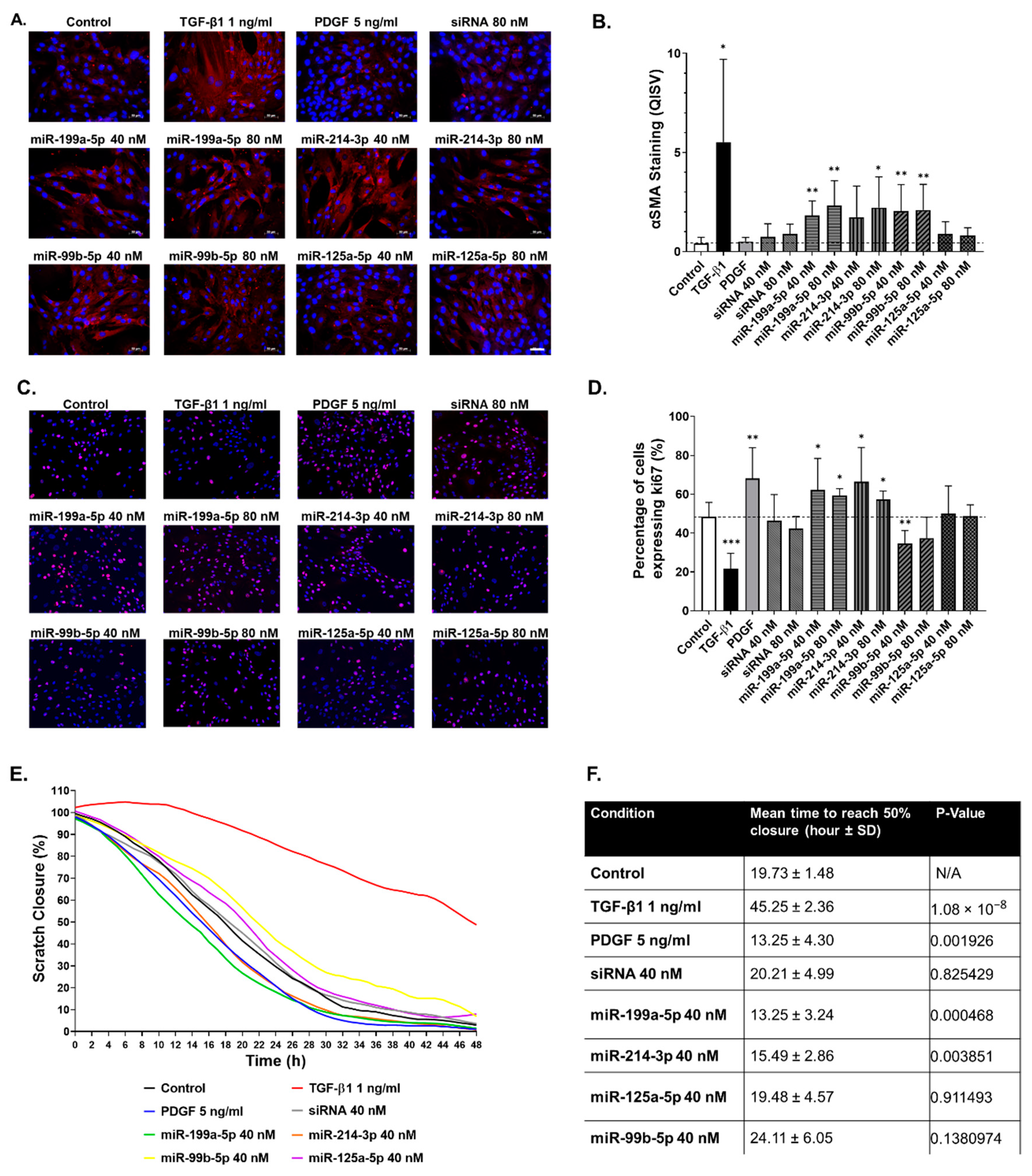
2.5. Functional Impact of Selected miRNAs on hTERT-HSC
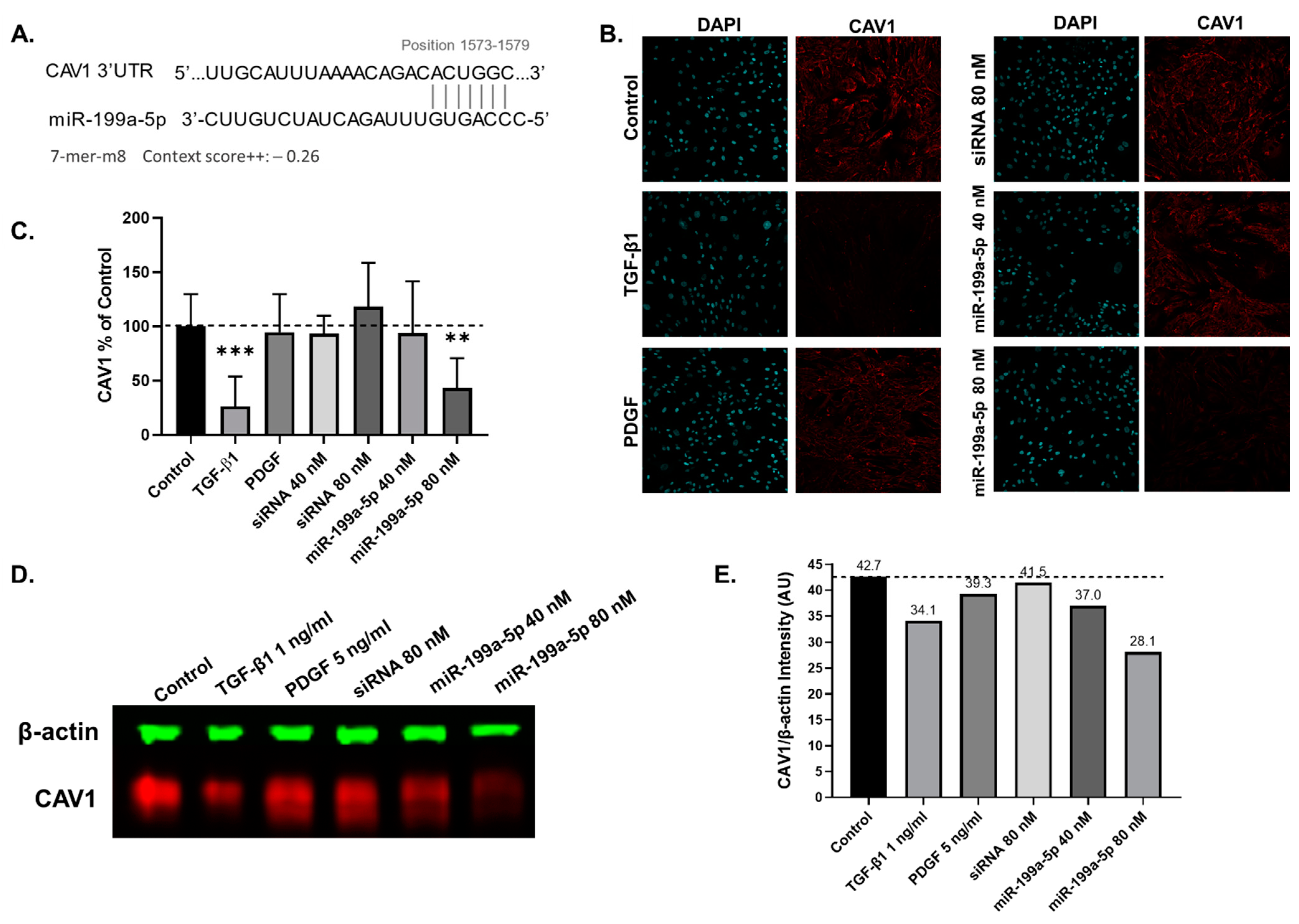
2.6. miR-199a-5p Targets Caveolin-1 in hTERT-HSCs
3. Discussion
3.1. Characterisation of Human Liver MTs
3.2. Compound-Specific Response of MTs Exposed to MTX and APAP
3.3. Extracellular miRNAs as Potential Markers of Fibrosis
3.4. miR-199a-5p, miR-214-3p and miR-99b-5p Promote hTERT-HSC Activation
4. Materials and Methods
4.1. Human Cell Lines
4.2. Generation of Microtissues
4.3. Induction of CYP3A4
4.4. Cytochrome P450 Assay
4.5. HPLC-MS/MS
4.6. Cell Treatments
4.7. Cell Viability Assay
4.8. Gene Expression Analysis
4.9. Next-Generation Sequencing
4.10. miRNA Analysis
4.11. Immunohistochemistry
4.12. Enzyme-Linked Immunosorbent Assay
4.13. Transfection of miRNA Mimics
4.14. Migration/Wound Healing Assay
4.15. Western Blot
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| ALT | Alanine aminotransferase |
| AOP | Adverse outcome pathway |
| APAP | Acetaminophen |
| AST | Aspartate aminotransferase |
| CAV1 | Caveolin-1 |
| CAV2 | Caveolin-2 |
| Col 4 | Collagen IV alpha 1 |
| Col I | Collagen I alpha 1 |
| CYP | Cytochrome P450 |
| ECM | Extracellular matrix |
| FBS | Fetal bovine serum |
| FN1 | Fibronectin |
| GSH | Glutathione |
| HSC | Hepatic stellate cell |
| hTERT-HSC | Immortal activated human hepatic stellate cells |
| miRNA | Micro RNA |
| mRNA | Messenger RNA |
| MTs | Microtissues |
| MTX | Methotrexate |
| NPC | Non-parenchymal cells |
| PFA | Paraformaldehyde |
| PHH | Primary human hepatocytes |
| PHMTs | Primary human microtissues |
| PMA | Phorbol 12-myristate 12-acetate |
| RIF | Rifampicin |
| TAA | Thioacetamide |
| TBL | Total bilirubin |
| TGF-β1 | Transforming growth factor-β |
| TNF-α | Tumour necrosis factor alpha |
| UMI | Unique Molecular Identifiers |
| αSMA | α-smooth muscle actin |
References
- Horvat, T.; Landesmann, B.; Lostia, A.; Vinken, M.; Munn, S.; Whelan, M. Adverse Outcome Pathway Development from Protein Alkylation to Liver Fibrosis. Arch. Toxicol. 2017, 91, 1523–1543. [Google Scholar] [CrossRef] [Green Version]
- Bataller, R.; Brenner, D.A. Liver Fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- BENYON, R.; IREDALE, J. Is Liver Fibrosis Reversible? Gut 2000, 46, 443–446. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, J.M.; Sagar, V.M.; Shah, T.; Shetty, S. Carcinogenesis on the Background of Liver Fibrosis: Implications for the Management of Hepatocellular Cancer. World J. Gastroenterol. 2018, 24, 4436–4447. [Google Scholar] [CrossRef] [PubMed]
- Zoubek, M.E.; Trautwein, C.; Strnad, P. Reversal of Liver Fibrosis: From Fiction to Reality. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 129–141. [Google Scholar] [CrossRef]
- Trautwein, C.; Friedman, S.L.; Schuppan, D.; Pinzani, M. Hepatic Fibrosis: Concept to Treatment. J. Hepatol. 2015, 62, S15–S24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepulveda-Crespo, D.; Resino, S.; Martinez, I. Strategies Targeting the Innate Immune Response for the Treatment of Hepatitis C Virus-Associated Liver Fibrosis. Drugs 2021, 81, 419–443. [Google Scholar] [CrossRef]
- David, S.; Hamilton, J.P. Drug-Induced Liver Injury. US Gastroenterol. Hepatol. Rev. 2010, 6, 73–80. [Google Scholar]
- Aithal, G.P.; Haugk, B.; Das, S.; Card, T.; Burt, A.D.; Record, C.O. Monitoring Methotrexate-Induced Hepatic Fibrosis in Patients with Psoriasis: Are Serial Liver Biopsies Justified? Aliment. Pharmacol. Ther. 2004, 19, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Berends, M.A.M.; van Oijen, M.G.H.; Snoek, J.; van de Kerkhof, P.C.M.; Drenth, J.P.H.; van Krieken, J.H.; de Jong, E.M.G.J. Reliability of the Roenigk Classification of Liver Damage After Methotrexate Treatment for Psoriasis: A Clinicopathologic Study of 160 Liver Biopsy Specimens. Arch. Dermatol. 2007, 143, 1515–1519. [Google Scholar] [CrossRef]
- Ramachandran, A.; Jaeschke, H. Mechanisms of Acetaminophen Hepatotoxicity and Their Translation to the Human Pathophysiology. J. Clin. Transl. Res. 2017, 3, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Alberti, A. Non Invasive Fibrosis Biomarkers Reduce but Not Substitute the Need for Liver Biopsy. World J. Gastroenterol. 2006, 12, 3682–3694. [Google Scholar] [CrossRef]
- Nallagangula, K.S.; Nagaraj, S.K.; Venkataswamy, L.; Chandrappa, M. Liver Fibrosis: A Compilation on the Biomarkers Status and Their Significance during Disease Progression. Future Sci. OA 2017, 4, FSO250. [Google Scholar] [CrossRef] [Green Version]
- Lurie, Y.; Webb, M.; Cytter-Kuint, R.; Shteingart, S.; Lederkremer, G.Z. Non-Invasive Diagnosis of Liver Fibrosis and Cirrhosis. World J. Gastroenterol. 2015, 21, 11567–11583. [Google Scholar] [CrossRef] [PubMed]
- Koda, M.; Matunaga, Y.; Kawakami, M.; Kishimoto, Y.; Suou, T.; Murawaki, Y. FibroIndex, a Practical Index for Predicting Significant Fibrosis in Patients with Chronic Hepatitis C. Hepatology 2007, 45, 297–306. [Google Scholar] [CrossRef]
- Allan, R.; Thoirs, K.; Phillips, M. Accuracy of Ultrasound to Identify Chronic Liver Disease. World J. Gastroenterol. 2010, 16, 3510–3520. [Google Scholar] [CrossRef] [PubMed]
- Arena, U.; Vizzutti, F.; Corti, G.; Ambu, S.; Stasi, C.; Bresci, S.; Moscarella, S.; Boddi, V.; Petrarca, A.; Laffi, G.; et al. Acute Viral Hepatitis Increases Liver Stiffness Values Measured by Transient Elastography. Hepatology 2008, 47, 380–384. [Google Scholar] [CrossRef]
- Bellan, M.; Castello, L.M.; Pirisi, M. Candidate Biomarkers of Liver Fibrosis: A Concise, Pathophysiology-Oriented Review. J. Clin. Transl. Hepatol. 2018, 6, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating MiRNA Panels for Specific and Early Detection in Bladder Cancer. Cancer Sci. 2019, 110, 408–419. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of MicroRNAs in Serum: A Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Noferesti, S.S.; Sohel, M.M.H.; Hoelker, M.; Salilew-Wondim, D.; Tholen, E.; Looft, C.; Rings, F.; Neuhoff, C.; Schellander, K.; Tesfaye, D. Controlled Ovarian Hyperstimulation Induced Changes in the Expression of Circulatory MiRNA in Bovine Follicular Fluid and Blood Plasma. J. Ovarian Res. 2015, 8, 81. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Starkey Lewis, P.J.; Dear, J.; Platt, V.; Simpson, K.J.; Craig, D.G.N.; Antoine, D.J.; French, N.S.; Dhaun, N.; Webb, D.J.; Costello, E.M.; et al. Circulating MicroRNAs as Potential Markers of Human Drug-Induced Liver Injury. Hepatology 2011, 54, 1767–1776. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.-Y. Secreted MicroRNAs: A New Form of Intercellular Communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted Monocytic MiR-150 Enhances Targeted Endothelial Cell Migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.; Zhang, L.; Xie, J.; Liu, F.; He, J.; He, J.; Wang, X.; Xiang, T. MicroRNA-146b Regulates Hepatic Stellate Cell Activation via Targeting of KLF4. Ann. Hepatol. 2016, 15, 918–928. [Google Scholar] [CrossRef]
- Murakami, Y.; Toyoda, H.; Tanaka, M.; Kuroda, M.; Harada, Y.; Matsuda, F.; Tajima, A.; Kosaka, N.; Ochiya, T.; Shimotohno, K. The Progression of Liver Fibrosis Is Related with Overexpression of the MiR-199 and 200 Families. PLoS ONE 2011, 6, e16081. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Enomoto, M.; Fujii, H.; Sekiya, Y.; Yoshizato, K.; Ikeda, K.; Kawada, N. MicroRNA-221/222 Upregulation Indicates the Activation of Stellate Cells and the Progression of Liver Fibrosis. Gut 2012, 61, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Markovic, J.; Sharma, A.D.; Balakrishnan, A. MicroRNA-221: A Fine Tuner and Potential Biomarker of Chronic Liver Injury. Cells 2020, 9, 1767. [Google Scholar] [CrossRef]
- Krauskopf, J.; de Kok, T.M.; Schomaker, S.J.; Gosink, M.; Burt, D.A.; Chandler, P.; Warner, R.L.; Johnson, K.J.; Caiment, F.; Kleinjans, J.C.; et al. Serum MicroRNA Signatures as “Liquid Biopsies” for Interrogating Hepatotoxic Mechanisms and Liver Pathogenesis in Human. PLoS ONE 2017, 12, e0177928. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Sun, C.; Zhao, F.; Zhang, Y.; Li, D. MiR-33a Levels in Hepatic and Serum after Chronic HBV-Induced Fibrosis. J. Gastroenterol. 2015, 50, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Lendvai, G.; Jármay, K.; Karácsony, G.; Halász, T.; Kovalszky, I.; Baghy, K.; Wittmann, T.; Schaff, Z.; Kiss, A. Elevated MiR-33a and MiR-224 in Steatotic Chronic Hepatitis C Liver Biopsies. World J. Gastroenterol. 2014, 20, 15343–15350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Wu, C.; Xu, Z.; Xia, P.; Dong, P.; Chen, B.; Yu, F. Hepatic Stellate Cell Is Activated by MicroRNA-181b via PTEN/Akt Pathway. Mol. Cell Biochem. 2015, 398, 1–9. [Google Scholar] [CrossRef]
- Wang, B.; Li, W.; Guo, K.; Xiao, Y.; Wang, Y.; Fan, J. MiR-181b Promotes Hepatic Stellate Cells Proliferation by Targeting P27 and Is Elevated in the Serum of Cirrhosis Patients. Biochem. Biophys. Res. Commun. 2012, 421, 4–8. [Google Scholar] [CrossRef]
- Yu, F.; Guo, Y.; Chen, B.; Dong, P.; Zheng, J. MicroRNA-17-5p Activates Hepatic Stellate Cells through Targeting of Smad7. Lab. Investig. 2015, 95, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of MiRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waidmann, O.; Bihrer, V.; Pleli, T.; Farnik, H.; Berger, A.; Zeuzem, S.; Kronenberger, B.; Piiper, A. Serum MicroRNA-122 Levels in Different Groups of Patients with Chronic Hepatitis B Virus Infection. J. Viral Hepat. 2012, 19, e58–e65. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between Circulating MicroRNAs (MiR-21, MiR-34a, MiR-122 and MiR-451) and Non-Alcoholic Fatty Liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Howell, L.S.; Ireland, L.; Park, B.K.; Goldring, C.E. MiR-122 and Other MicroRNAs as Potential Circulating Biomarkers of Drug-Induced Liver Injury. Expert Rev. Mol. Diagn. 2018, 18, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kia, R.; Kelly, L.; Sison-Young, R.L.C.; Zhang, F.; Pridgeon, C.S.; Heslop, J.A.; Metcalfe, P.; Kitteringham, N.R.; Baxter, M.; Harrison, S.; et al. MicroRNA-122: A Novel Hepatocyte-Enriched in Vitro Marker of Drug-Induced Cellular Toxicity. Toxicol. Sci. 2015, 144, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Carroll, E.E.; Ippolito, D.L.; Permenter, M.G.; McDyre, B.C.; Baer, C.E.; Kumsher, D.M.; Boyle, M.H.; DiVito, V.T.; Lewis, J.A.; Koontz, J.M. Utility of Serum MiR-122, Liver Enzymes, and Hepatic Histopathology in Response to Hepatotoxicants in Sprague-Dawley Rats. Toxicol. Pathol. 2018, 46, 835–846. [Google Scholar] [CrossRef]
- Messner, C.J.; Premand, C.; Gaiser, C.; Kluser, T.; Kübler, E.; Suter-Dick, L. Exosomal MicroRNAs Release as a Sensitive Marker for Drug-Induced Liver Injury In Vitro. Appl. In Vitro Toxicol. 2020, 6, 77–89. [Google Scholar] [CrossRef]
- Norona, L.M.; Nguyen, D.G.; Gerber, D.A.; Presnell, S.C.; Mosedale, M.; Watkins, P.B. Bioprinted Liver Provides Early Insight into the Role of Kupffer Cells in TGF-Β1 and Methotrexate-Induced Fibrogenesis. PLoS ONE 2019, 14, e0208958. [Google Scholar] [CrossRef]
- Devaraj Ezhilarasan Role of MicroRNAs in Hepatic Fibrosis Progression. J. Appl. Pharm. Sci. 2018. [CrossRef] [Green Version]
- Loosen, S.H.; Schueller, F.; Trautwein, C.; Roy, S.; Roderburg, C. Role of Circulating MicroRNAs in Liver Diseases. World J. Hepatol. 2017, 9, 586–594. [Google Scholar] [CrossRef]
- Iacob, D.G.; Rosca, A.; Ruta, S.M. Circulating MicroRNAs as Non-Invasive Biomarkers for Hepatitis B Virus Liver Fibrosis. World J. Gastroenterol. 2020, 26, 1113. [Google Scholar] [CrossRef]
- Prestigiacomo, V.; Weston, A.; Messner, S.; Lampart, F.; Suter-Dick, L. Pro-Fibrotic Compounds Induce Stellate Cell Activation, ECM-Remodelling and Nrf2 Activation in a Human 3D-Multicellular Model of Liver Fibrosis. PLoS ONE 2017, 12, e0179995. [Google Scholar] [CrossRef] [PubMed]
- Lino Cardenas, C.L.; Henaoui, I.S.; Courcot, E.; Roderburg, C.; Cauffiez, C.; Aubert, S.; Copin, M.-C.; Wallaert, B.; Glowacki, F.; Dewaeles, E.; et al. MiR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1. PLoS Genet. 2013, 9, e1003291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Ma, L.; Wei, R.; Ye, T.; Zhou, J.; Wen, M.; Men, R.; Aqeilan, R.I.; Peng, Y.; Yang, L. Twist1-Induced MiR-199a-3p Promotes Liver Fibrosis by Suppressing Caveolin-2 and Activating TGF-β Pathway. Signal Transduct. Target. Ther. 2020, 5, 1–12. [Google Scholar] [CrossRef]
- Roy, S.; Benz, F.; Luedde, T.; Roderburg, C. The Role of MiRNAs in the Regulation of Inflammatory Processes during Hepatofibrogenesis. Hepatobiliary Surg. Nutr. 2015, 4, 24–33. [Google Scholar] [CrossRef]
- Ma, L.; Yang, X.; Wei, R.; Ye, T.; Zhou, J.-K.; Wen, M.; Men, R.; Li, P.; Dong, B.; Liu, L.; et al. MicroRNA-214 Promotes Hepatic Stellate Cell Activation and Liver Fibrosis by Suppressing Sufu Expression. Cell Death Dis. 2018, 9, 718. [Google Scholar] [CrossRef]
- Coppola, N.; Onorato, L.; Panella, M.; de Stefano, G.; Mosca, N.; Minichini, C.; Messina, V.; Potenza, N.; Starace, M.; Alessio, L.; et al. Correlation Between the Hepatic Expression of Human MicroRNA Hsa-MiR-125a-5p and the Progression of Fibrosis in Patients With Overt and Occult HBV Infection. Front. Immunol. 2018, 9, 1334. [Google Scholar] [CrossRef] [Green Version]
- Oura, K.; Fujita, K.; Morishita, A.; Iwama, H.; Nakahara, M.; Tadokoro, T.; Sakamoto, T.; Nomura, T.; Yoneyama, H.; Mimura, S.; et al. Serum MicroRNA-125a-5p as a Potential Biomarker of HCV-associated Hepatocellular Carcinoma. Oncol. Lett. 2019, 18, 882–890. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Li, J.; Li, C.; Qi, H.; Dong, P.; Zheng, J.; Yu, F. MicroRNA-125a-5p Contributes to Hepatic Stellate Cell Activation through Targeting FIH1. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 38, 1544–1552. [Google Scholar] [CrossRef]
- Estep, M.; Armistead, D.; Hossain, N.; Elarainy, H.; Goodman, Z.; Baranova, A.; Chandhoke, V.; Younossi, Z.M. Differential Expression of MiRNAs in the Visceral Adipose Tissue of Patients with Non-Alcoholic Fatty Liver Disease. Aliment. Pharmacol. Ther. 2010, 32, 487–497. [Google Scholar] [CrossRef]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 Family Targets AKT/MTOR Signaling Pathway in Dermal Wound Healing. PLoS ONE 2013, 8, e64434. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Liu, T.; Sun, S.; Feng, S. The Role of the MiR-99b-5p/MTOR Signaling Pathway in Neuroregeneration in Mice Following Spinal Cord Injury. Mol. Med. Rep. 2017, 16, 9355–9360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Chan, E.; Li, X.; Huang, M. Clinical Outcomes and Management of Mechanism-Based Inhibition of Cytochrome P450 3A4. Ther. Clin. Risk Manag. 2005, 1, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueyama, T.; Tsuji, S.; Sugiyama, T.; Tada, M. Fluorometric Evaluation of CYP3A4 Expression Using Improved Transgenic HepaRG Cells Carrying a Dual-Colour Reporter for CYP3A4 and CYP3A7. Sci. Rep. 2017, 7, 2874. [Google Scholar] [CrossRef]
- Berger, B.; Donzelli, M.; Maseneni, S.; Boess, F.; Roth, A.; Krähenbühl, S.; Haschke, M. Comparison Of Liver Cell Models Using The Basel Phenotyping Cocktail. Front. Pharmacol. 2016, 7, 443. [Google Scholar] [CrossRef] [Green Version]
- Mandon, M.; Huet, S.; Dubreil, E.; Fessard, V.; Hégarat, L.L. Three-Dimensional HepaRG Spheroids as a Liver Model to Study Human Genotoxicity in Vitro with the Single Cell Gel Electrophoresis Assay. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Yan, L.; Messner, C.J.; Zhang, X.; Suter-Dick, L. Assessment of Fibrotic Pathways Induced by Environmental Chemicals Using 3D-Human Liver Microtissue Model. Environ. Res. 2021, 194, 110679. [Google Scholar] [CrossRef]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D Organotypic Cultures of Human HepaRG Cells: A Tool for In Vitro Toxicity Studies. Toxicol. Sci. 2013, 133, 67–78. [Google Scholar] [CrossRef]
- Levitt, D.G.; Levitt, M.D. Human Serum Albumin Homeostasis: A New Look at the Roles of Synthesis, Catabolism, Renal and Gastrointestinal Excretion, and the Clinical Value of Serum Albumin Measurements. Int. J. Gen. Med. 2016, 9, 229–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lübberstedt, M.; Müller-Vieira, U.; Mayer, M.; Biemel, K.M.; Knöspel, F.; Knobeloch, D.; Nüssler, A.K.; Gerlach, J.C.; Zeilinger, K. HepaRG Human Hepatic Cell Line Utility as a Surrogate for Primary Human Hepatocytes in Drug Metabolism Assessment in Vitro. J. Pharmacol. Toxicol. Methods 2011, 63, 59–68. [Google Scholar] [CrossRef]
- Prestigiacomo, V.; Suter-Dick, L. Nrf2 Protects Stellate Cells from Smad-Dependent Cell Activation. PLoS ONE 2018, 13, e0201044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, C.C.; Dankers, A.C.A.; Lauschke, V.M.; Sison-Young, R.; Jenkins, R.; Rowe, C.; Goldring, C.E.; Park, K.; Regan, S.L.; Walker, T.; et al. Comparison of Hepatic 2D Sandwich Cultures and 3D Spheroids for Long-Term Toxicity Applications: A Multicenter Study. Toxicol. Sci. 2018, 162, 655–666. [Google Scholar] [CrossRef] [Green Version]
- Hinson, J.A.; Roberts, D.W.; James, L.P. Mechanisms of Acetaminophen-Induced Liver Necrosis. Handb. Exp. Pharmacol. 2010, 369–405. [Google Scholar] [CrossRef] [Green Version]
- Leite, S.B.; Roosens, T.; El Taghdouini, A.; Mannaerts, I.; Smout, A.J.; Najimi, M.; Sokal, E.; Noor, F.; Chesne, C.; van Grunsven, L.A. Novel Human Hepatic Organoid Model Enables Testing of Drug-Induced Liver Fibrosis In Vitro. Biomaterials 2016, 78, 1–10. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.; Choi, J.; Jung, H.; Lee, K.; Kang, J.; Park, N.; Rim, Y.A.; Nam, Y.; Ju, J.H. Recapitulation of Methotrexate Hepatotoxicity with Induced Pluripotent Stem Cell-Derived Hepatocytes from Patients with Rheumatoid Arthritis. Stem Cell Res. Ther. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Barker, J.; Horn, E.J.; Lebwohl, M.; Warren, R.B.; Nast, A.; Rosenberg, W.; Smith, C. International Psoriasis Council Assessment and Management of Methotrexate Hepatotoxicity in Psoriasis Patients: Report from a Consensus Conference to Evaluate Current Practice and Identify Key Questions toward Optimizing Methotrexate Use in the Clinic. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 758–764. [Google Scholar] [CrossRef]
- Mungunsukh, O.; Day, R.M. Transforming Growth Factor-Β1 Selectively Inhibits Hepatocyte Growth Factor Expression via a Micro-RNA-199-Dependent Posttranscriptional Mechanism. Mol. Biol. Cell 2013, 24, 2088–2097. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, M.; Ogawa, T.; Enomoto, M.; Motoyama, H.; Yoshizato, K.; Ikeda, K.; Kawada, N. Induction of MicroRNA-214-5p in Human and Rodent Liver Fibrosis. Fibrogenesis Tissue Repair 2012, 5, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Reilly, S. MicroRNAs in Fibrosis: Opportunities and Challenges. Arthritis Res. Ther. 2016, 18, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Ye, B.; Wang, P.; Yao, F.; Zhang, C.; Yu, G. Overview of MicroRNA-199a Regulation in Cancer. Cancer Manag. Res. 2019, 11, 10327–10335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Huang, B.; Wang, H.-Y.; Chang, A.; Zheng, X.F.S. Emerging Role of MicroRNAs in MTOR Signaling. Cell. Mol. Life Sci. 2017, 74, 2613–2625. [Google Scholar] [CrossRef]
- Turcatel, G.; Rubin, N.; El-Hashash, A.; Warburton, D. MIR-99a and MIR-99b Modulate TGF-β Induced Epithelial to Mesenchymal Plasticity in Normal Murine Mammary Gland Cells. PLoS ONE 2012, 7, e31032. [Google Scholar] [CrossRef]
- Wang, G.; Mao, W.; Zheng, S.; Ye, J. Epidermal Growth Factor Receptor-Regulated MiR-125a-5p—A Metastatic Inhibitor of Lung Cancer. FEBS J. 2009, 276, 5571–5578. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Sun, S.; Shi, J.; Cao, F.; Han, X.; Chen, Z. MicroRNA-125a-5p Plays a Role as a Tumor Suppressor in Lung Carcinoma Cells by Directly Targeting STAT3. Tumour Biol. 2017, 39, 1010428317697579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| miRNA | In Vitro | In Vivo | Clinical | Publications |
|---|---|---|---|---|
| miR-199a-5p |
|
|
| Cardenas et al., 2013 [50] Yang et al., 2020 [51] Ezhilarasan, 2018 [46] Roy et al., 2015 [52] |
| miR-214-3p |
|
|
| Ma et al., 2018 [53] Ezhilarasan, 2018 [46] Krauskopf et al., 2017 [32] |
| miR-125a-5p |
|
|
| Coppola et al., 2018 [54] Oura et al., 2019 [55] Li et al., 2016 [56] |
| miR-99b-5p |
|
|
| Estep et al., 2010 [57] Jin et al., 2013 [58] Cao et al., 2017 [59] |
| Plate Size | Cell Number | Concentration (mM) | HiPerfect Reagent | Mimic | DMEM without Supplements | Final Medium + Reagent Quantity per Well |
|---|---|---|---|---|---|---|
| 96-well | 2000 | 40 | 0.75 µL | 0.12 µL | 24.25 µL | 200 µL |
| 80 | 0.75 µL | 0.24 µL | 24.25 µL | |||
| 48-well | 10,000 | 40 | 1.5 µL | 0.24 µL | 48.5 µL | 400 µL |
| 80 | 1.5 µL | 0.48 µL | 48.5 µL | |||
| 6-well | 60,000 | 40 | 6 µL | 0.96 µL | 194 µL | 1600 µL |
| 80 | 6 µL | 1.92 µL | 194 µL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messner, C.J.; Schmidt, S.; Özkul, D.; Gaiser, C.; Terracciano, L.; Krähenbühl, S.; Suter-Dick, L. Identification of miR-199a-5p, miR-214-3p and miR-99b-5p as Fibrosis-Specific Extracellular Biomarkers and Promoters of HSC Activation. Int. J. Mol. Sci. 2021, 22, 9799. https://doi.org/10.3390/ijms22189799
Messner CJ, Schmidt S, Özkul D, Gaiser C, Terracciano L, Krähenbühl S, Suter-Dick L. Identification of miR-199a-5p, miR-214-3p and miR-99b-5p as Fibrosis-Specific Extracellular Biomarkers and Promoters of HSC Activation. International Journal of Molecular Sciences. 2021; 22(18):9799. https://doi.org/10.3390/ijms22189799
Chicago/Turabian StyleMessner, Catherine Jane, Saskia Schmidt, Dilek Özkul, Carine Gaiser, Luigi Terracciano, Stephan Krähenbühl, and Laura Suter-Dick. 2021. "Identification of miR-199a-5p, miR-214-3p and miR-99b-5p as Fibrosis-Specific Extracellular Biomarkers and Promoters of HSC Activation" International Journal of Molecular Sciences 22, no. 18: 9799. https://doi.org/10.3390/ijms22189799






