Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor
Abstract
1. Introduction
2. Results
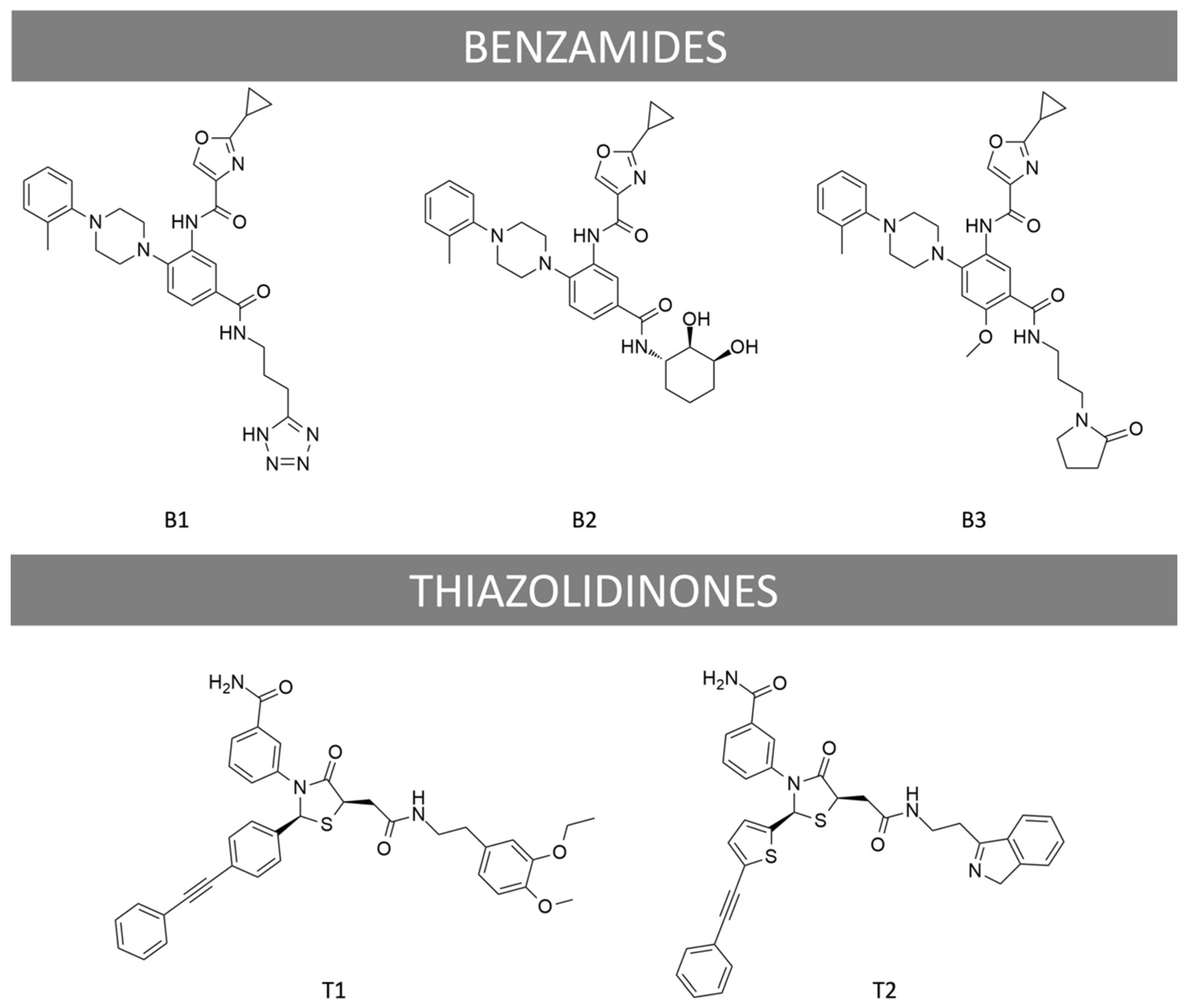
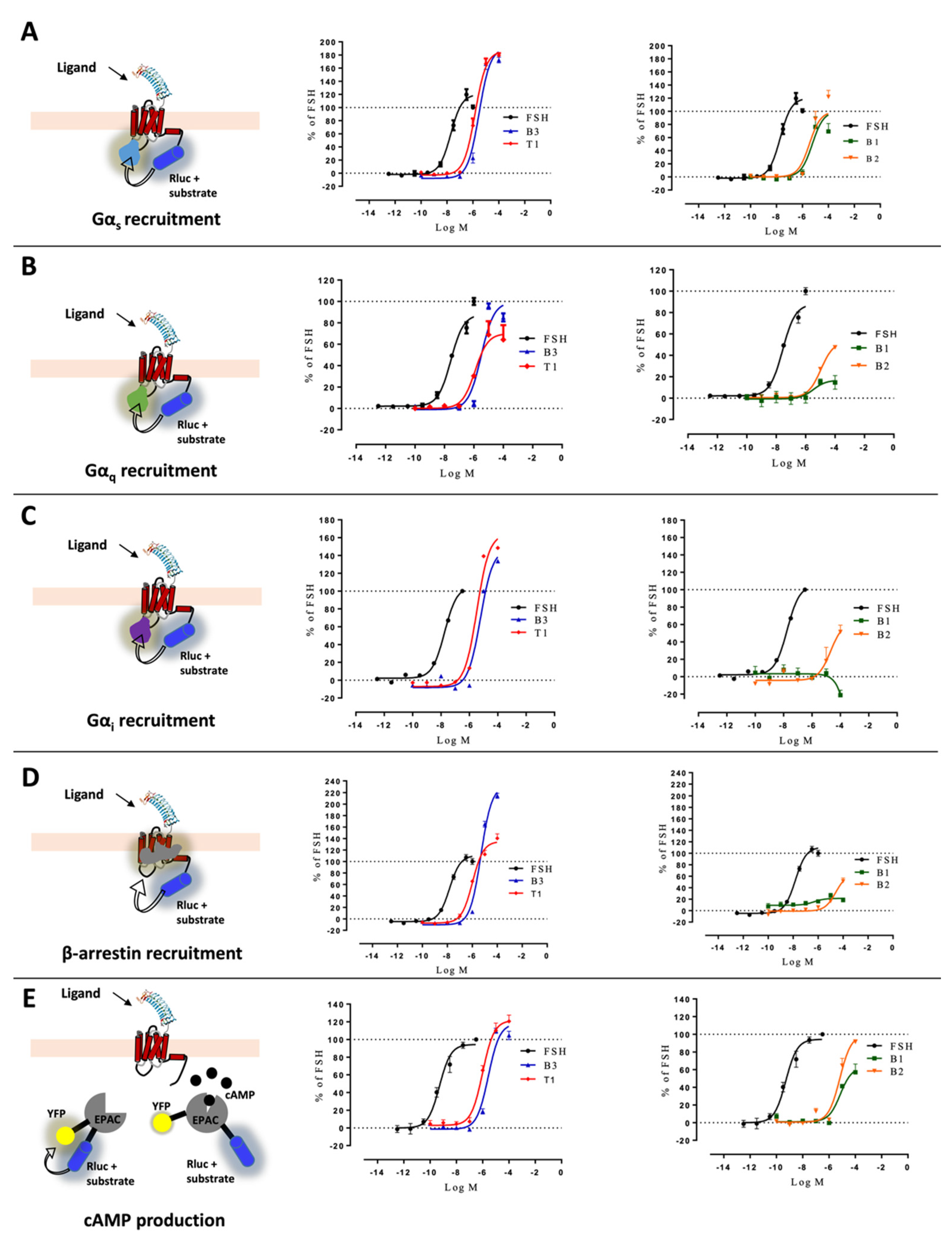
2.1. Diversity in the Recruitment of G Proteins, β-Arrestin 2, and in cAMP Production
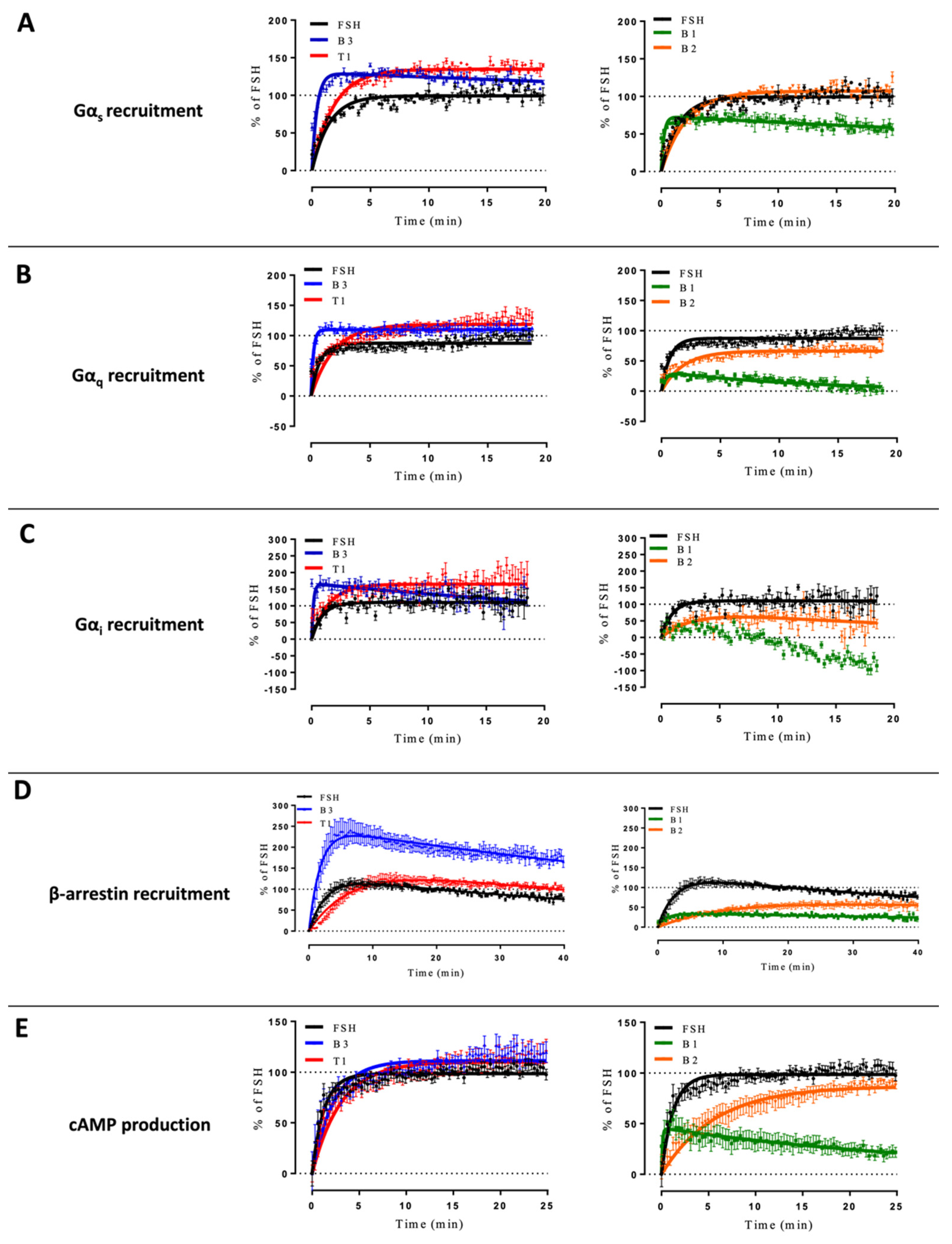
2.2. Differences in Kinetics That Affect Potencies and Efficacies
2.3. Diversity of FSHR-Mediated ERK Phosphorylation and cAMP-Responsive Element-Dependent Transcription
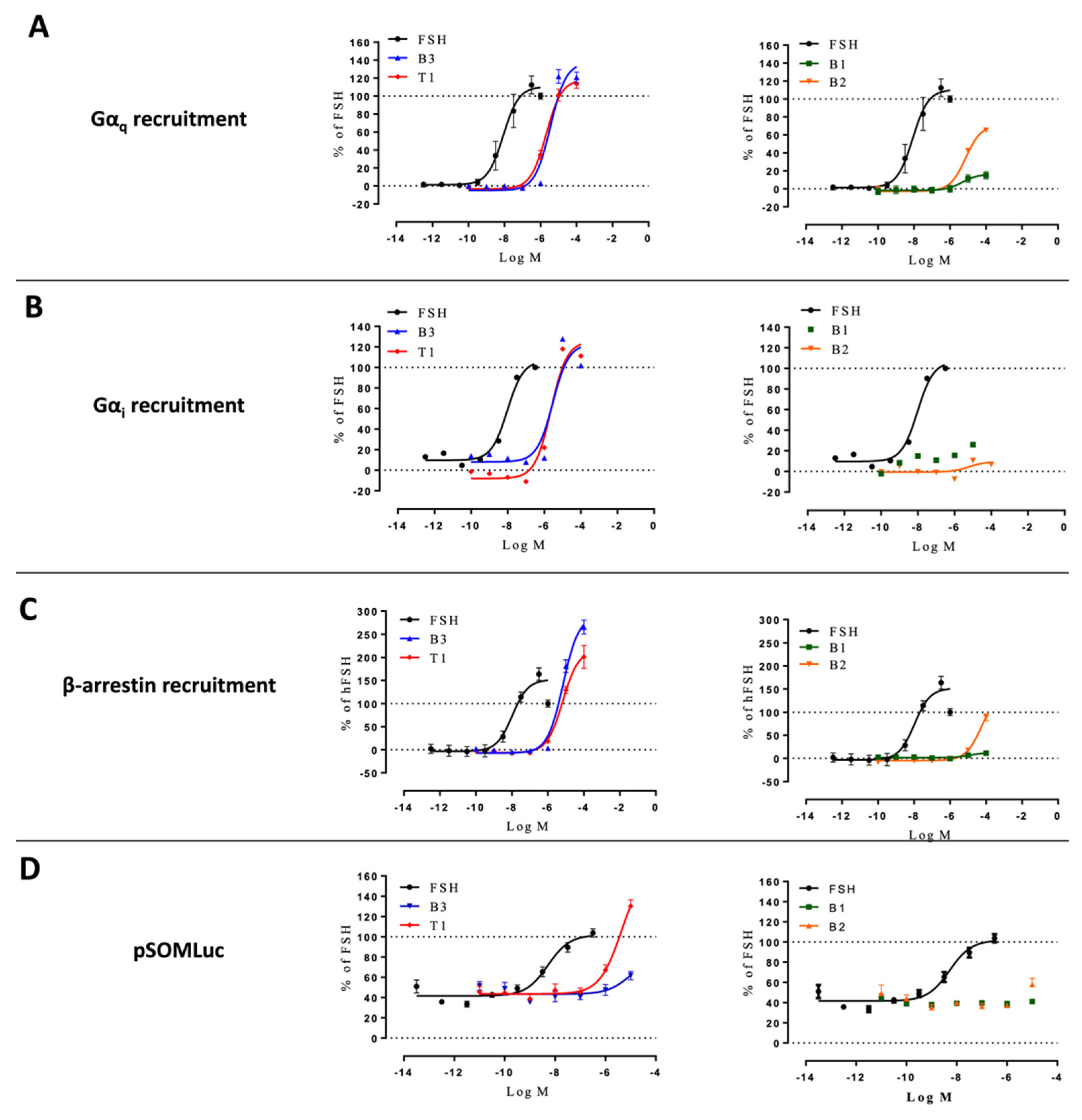
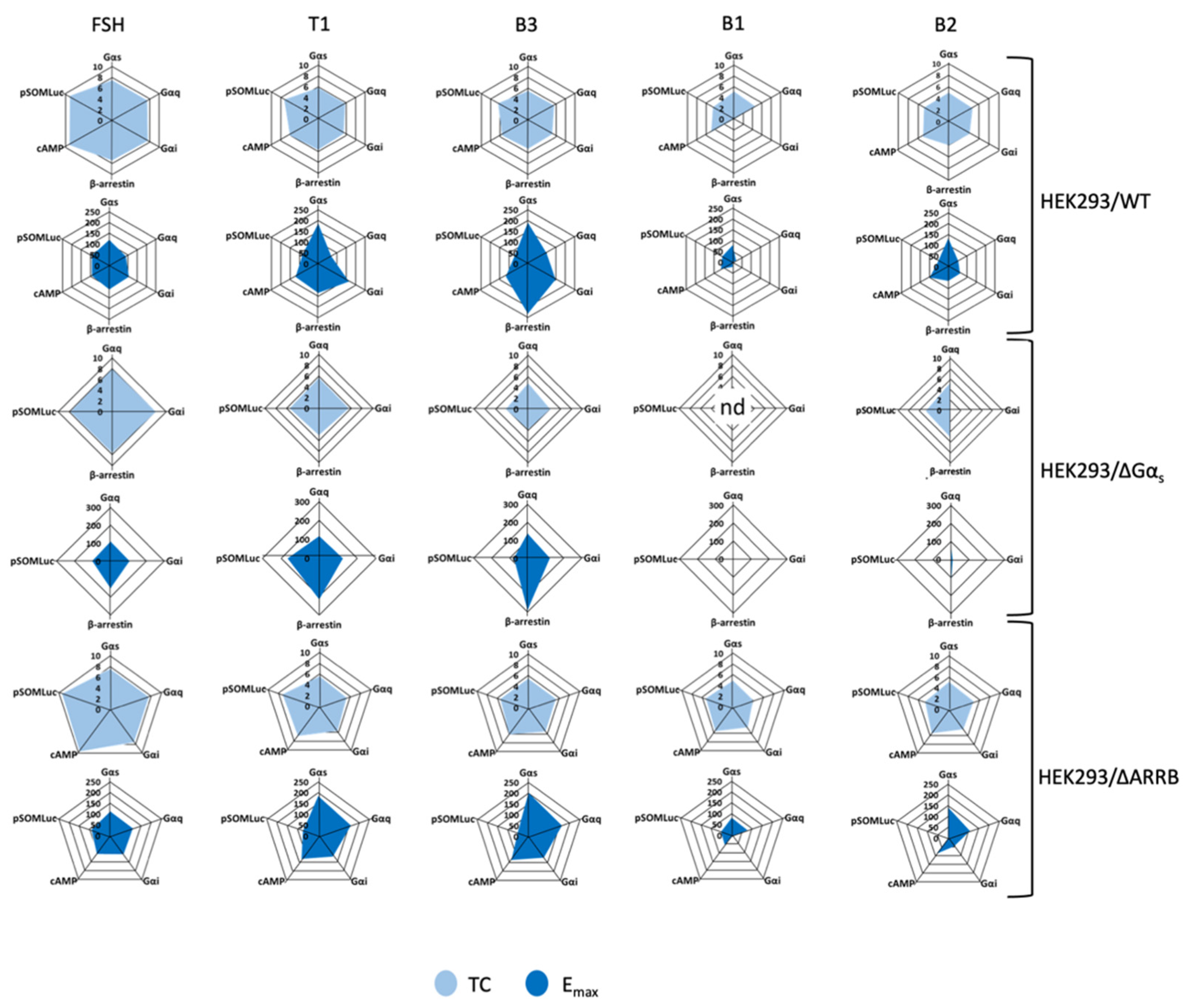
2.4. Profiling in HEK293/ΔGαs and HEK293/ΔARRB Cells
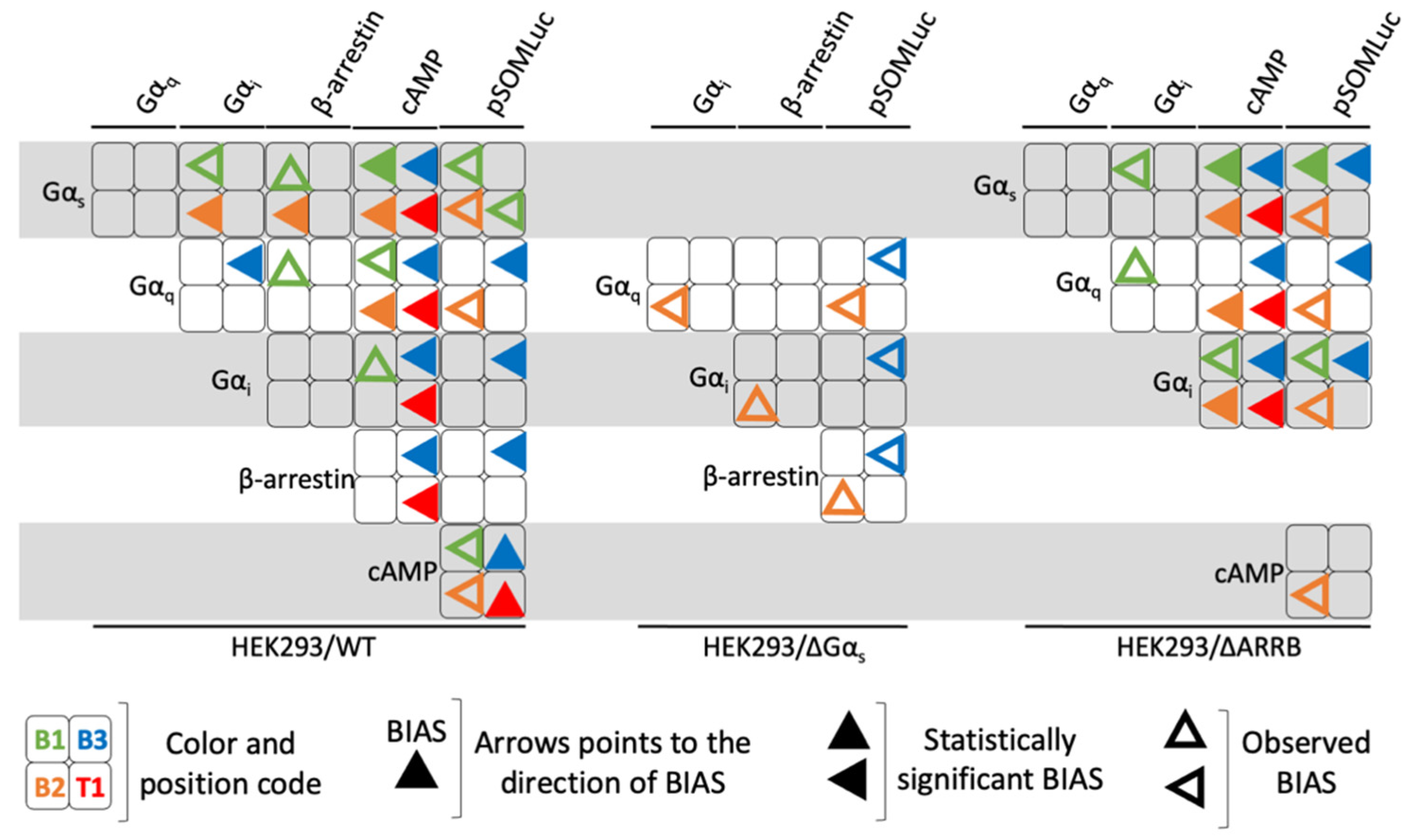
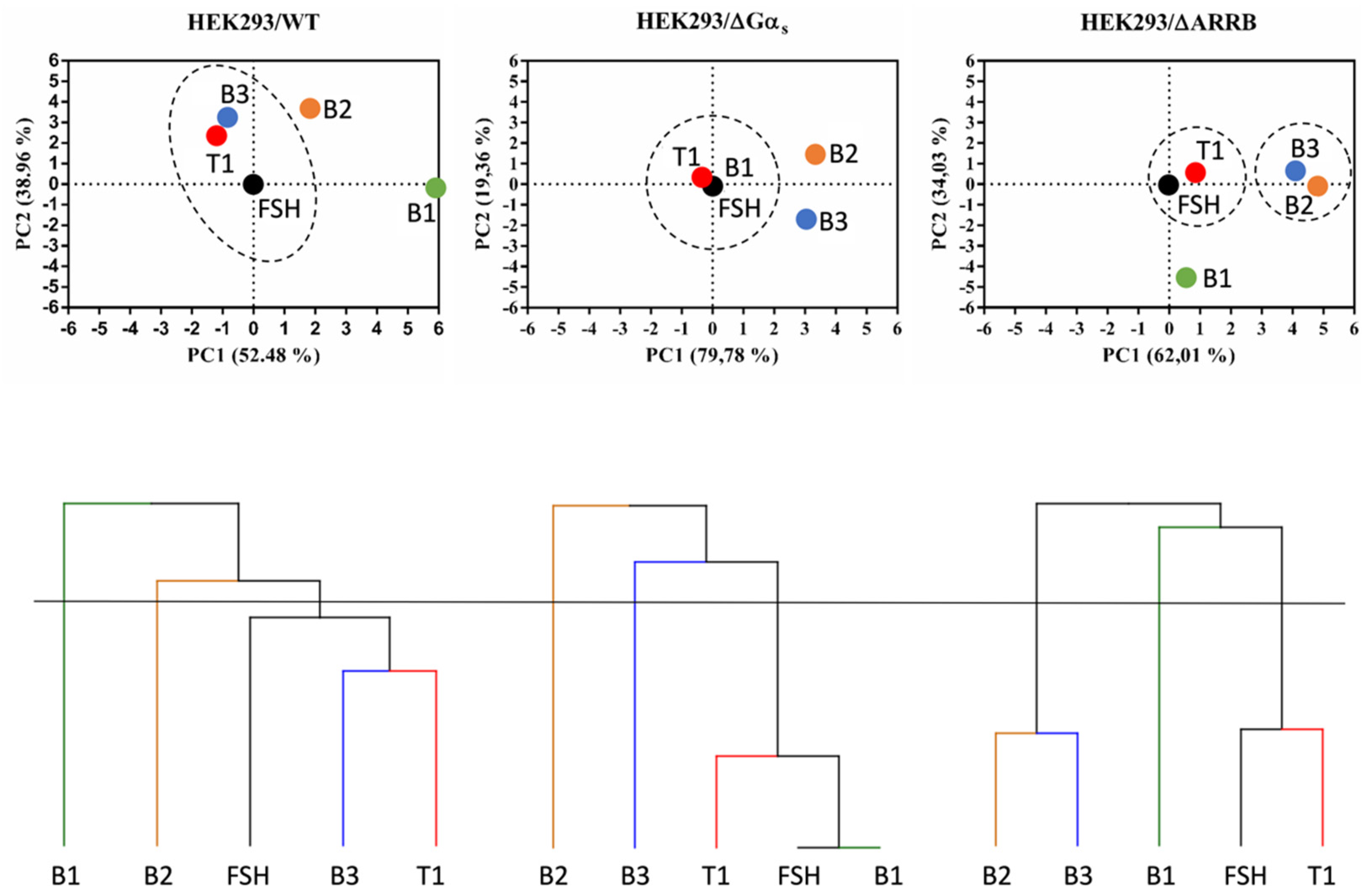
2.5. Analysis of Bias Factors and Multi-Dimensional Comparison of Pharmacological Efficacies
3. Discussion
4. Materials and Methods
4.1. Ligands and Materials
4.2. Cell Culture and Transfection
4.3. BRET Sensors
4.4. Cell Stimulation and BRET Measurement
4.5. Western Blot Analysis
4.6. Flow Cytometry
4.7. Cre-Dependent Reporter Assay
4.8. Statistical Analysis of Concentration/Activity Curves
4.9. Statistical Analysis of Kinetic Curves
4.10. Bias Calculation and Statistics
4.11. Dealing with Parameter Non-Identifiability in Bias Calculations
4.12. Principal Component Analysis (PCA) and Hierarchical Clustering
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gromoll, J.; Simoni, M.; Nordhoff, V.; Behre, H.M.; De Geyter, C.; Nieschlag, E. Functional and Clinical Consequences of Mutations in the FSH Receptor. Mol. Cell. Endocrinol. 1996, 125, 177–182. [Google Scholar] [CrossRef]
- Kaprara, A.; Huhtaniemi, I.T. The Hypothalamus-Pituitary-Gonad Axis: Tales of Mice and Men. Metabolism 2017, 86, 3–17. [Google Scholar] [CrossRef]
- Fan, Q.R.; Hendrickson, W.A. Structure of Human Follicle-Stimulating Hormone in Complex with Its Receptor. Nature 2005, 433, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Gloaguen, P.; Crépieux, P.; Heitzler, D.; Poupon, A.; Reiter, E. Mapping the Follicle-Stimulating Hormone-Induced Signaling Networks. Front. Endocrinol. 2011, 2, 45. [Google Scholar] [CrossRef] [PubMed]
- Landomiel, F.; De Pascali, F.; Raynaud, P.; Jean-Alphonse, F.; Yvinec, R.; Pellissier, L.P.; Bozon, V.; Bruneau, G.; Crépieux, P.; Poupon, A.; et al. Biased Signaling and Allosteric Modulation at the FSHR. Front. Endocrinol. 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Dattatreyamurty, B.; Figgs, L.W.; Reichert, L.E. Physical and Functional Association of Follitropin Receptors with Cholera Toxin-Sensitive Guanine Nucleotide-Binding Protein. J. Biol. Chem. 1987, 262, 11737–11745. [Google Scholar] [CrossRef]
- Salvador, L.M.; Park, Y.; Cottom, J.; Maizels, E.T.; Jones, J.C.; Schillace, R.V.; Carr, D.W.; Cheung, P.; Allis, C.D.; Jameson, J.L.; et al. Follicle-Stimulating Hormone Stimulates Protein Kinase A-Mediated Histone H3 Phosphorylation and Acetylation Leading to Select Gene Activation in Ovarian Granulosa Cells. J. Biol. Chem. 2001, 276, 40146–40155. [Google Scholar] [CrossRef] [PubMed]
- Cottom, J.; Salvador, L.M.; Maizels, E.T.; Reierstad, S.; Park, Y.; Carr, D.W.; Davare, M.A.; Hell, J.W.; Palmer, S.S.; Dent, P.; et al. Follicle-Stimulating Hormone Activates Extracellular Signal-Regulated Kinase but Not Extracellular Signal-Regulated Kinase Kinase through a 100-KDa Phosphotyrosine Phosphatase. J. Biol. Chem. 2003, 278, 7167–7179. [Google Scholar] [CrossRef] [PubMed]
- De Pascali, F.; Reiter, E. β-Arrestins and Biased Signaling in Gonadotropin Receptors. Minerva. Ginecol. 2018, 70, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Kara, E.; Crépieux, P.; Gauthier, C.; Martinat, N.; Piketty, V.; Guillou, F.; Reiter, E. A Phosphorylation Cluster of Five Serine and Threonine Residues in the C-Terminus of the Follicle-Stimulating Hormone Receptor Is Important for Desensitization but Not for Beta-Arrestin-Mediated ERK Activation. Mol. Endocrinol. 2006, 20, 3014–3026. [Google Scholar] [CrossRef]
- Tranchant, T.; Durand, G.; Gauthier, C.; Crépieux, P.; Ulloa-Aguirre, A.; Royère, D.; Reiter, E. Preferential β-Arrestin Signalling at Low Receptor Density Revealed by Functional Characterization of the Human FSH Receptor A189 V Mutation. Mol. Cell. Endocrinol. 2011, 331, 109–118. [Google Scholar] [CrossRef]
- Tréfier, A.; Musnier, A.; Landomiel, F.; Bourquard, T.; Boulo, T.; Ayoub, M.A.; León, K.; Bruneau, G.; Chevalier, M.; Durand, G.; et al. G Protein-Dependent Signaling Triggers a β-Arrestin-Scaffolded P70S6K/ RpS6 Module That Controls 5’TOP MRNA Translation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 1154–1169. [Google Scholar] [CrossRef]
- Okashah, N.; Wan, Q.; Ghosh, S.; Sandhu, M.; Inoue, A.; Vaidehi, N.; Lambert, N.A. Variable G Protein Determinants of GPCR Coupling Selectivity. Proc. Natl. Acad. Sci. USA 2019, 116, 12054–12059. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.; Hipkin, R.W.; Sánchez-Yagüe, J.; Ascoli, M. Follitropin (FSH) and a Phorbol Ester Stimulate the Phosphorylation of the FSH Receptor in Intact Cells. J. Biol. Chem. 1994, 269, 8772–8779. [Google Scholar] [CrossRef]
- Crépieux, P.; Marion, S.; Martinat, N.; Fafeur, V.; Vern, Y.L.; Kerboeuf, D.; Guillou, F.; Reiter, E. The ERK-Dependent Signalling Is Stage-Specifically Modulated by FSH, during Primary Sertoli Cell Maturation. Oncogene 2001, 20, 4696–4709. [Google Scholar] [CrossRef]
- Nataraja, S.; Sriraman, V.; Palmer, S. Allosteric Regulation of the Follicle-Stimulating Hormone Receptor. Endocrinology 2018, 159, 2704–2716. [Google Scholar] [CrossRef] [PubMed]
- Kobilka, B.K. Structural Insights into Adrenergic Receptor Function and Pharmacology. Trends Pharmacol. Sci. 2011, 32, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, R.; Zou, Y.; Dror, R.O.; Mildorf, T.J.; Arlow, D.H.; Manglik, A.; Pan, A.C.; Liu, C.W.; Fung, J.J.; Bokoch, M.P.; et al. The Dynamic Process of β(2)-Adrenergic Receptor Activation. Cell 2013, 152, 532–542. [Google Scholar] [CrossRef]
- Wacker, D.; Wang, C.; Katritch, V.; Han, G.W.; Huang, X.-P.; Vardy, E.; McCorvy, J.D.; Jiang, Y.; Chu, M.; Siu, F.Y.; et al. Structural Features for Functional Selectivity at Serotonin Receptors. Science 2013, 340, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Galandrin, S.; Oligny-Longpré, G.; Bouvier, M. The Evasive Nature of Drug Efficacy: Implications for Drug Discovery. Trends Pharmacol. Sci. 2007, 28, 423–430. [Google Scholar] [CrossRef]
- Kenakin, T. Ligand-Selective Receptor Conformations Revisited: The Promise and the Problem. Trends Pharmacol. Sci. 2003, 24, 346–354. [Google Scholar] [CrossRef]
- Violin, J.D.; Lefkowitz, R.J. Beta-Arrestin-Biased Ligands at Seven-Transmembrane Receptors. Trends Pharmacol. Sci. 2007, 28, 416–422. [Google Scholar] [CrossRef]
- Reiter, E.; Ahn, S.; Shukla, A.K.; Lefkowitz, R.J. Molecular Mechanism of β-Arrestin-Biased Agonism at Seven-Transmembrane Receptors. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 179–197. [Google Scholar] [CrossRef]
- Changeux, J.-P.; Christopoulos, A. Allosteric Modulation as a Unifying Mechanism for Receptor Function and Regulation. Cell 2016, 166, 1084–1102. [Google Scholar] [CrossRef]
- Wootten, D.; Christopoulos, A.; Sexton, P.M. Emerging Paradigms in GPCR Allostery: Implications for Drug Discovery. Nat. Rev. Drug Discov. 2013, 12, 630–644. [Google Scholar] [CrossRef]
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased Signalling: From Simple Switches to Allosteric Microprocessors. Nat. Rev. Drug Discov. 2018, 17, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Nataraja, S.G.; Yu, H.N.; Palmer, S.S. Discovery and Development of Small Molecule Allosteric Modulators of Glycoprotein Hormone Receptors. Front. Endocrinol. 2015, 6, 142. [Google Scholar] [CrossRef]
- Ulloa-Aguirre, A.; Reiter, E.; Crépieux, P. FSH Receptor Signaling: Complexity of Interactions and Signal Diversity. Endocrinology 2018, 159, 3020–3035. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.A.; Bonnet, B.; Weaver, B.A.; Watts, J.; Kluetzman, K.; Thomas, R.M.; Poli, S.; Mutel, V.; Campo, B. A Negative Allosteric Modulator Demonstrates Biased Antagonism of the Follicle Stimulating Hormone Receptor. Mol. Cell. Endocrinol. 2011, 333, 143–150. [Google Scholar] [CrossRef][Green Version]
- Dias, J.A.; Campo, B.; Weaver, B.A.; Watts, J.; Kluetzman, K.; Thomas, R.M.; Bonnet, B.; Mutel, V.; Poli, S.M. Inhibition of Follicle-Stimulating Hormone-Induced Preovulatory Follicles in Rats Treated with a Nonsteroidal Negative Allosteric Modulator of Follicle-Stimulating Hormone Receptor1. Biol. Reprod. 2014, 90, 1–11. [Google Scholar] [CrossRef]
- van Koppen, C.J.; Verbost, P.M.; van de Lagemaat, R.; Karstens, W.-J.F.; Loozen, H.J.J.; van Achterberg, T.A.E.; van Amstel, M.G.A.; Brands, J.H.G.M.; van Doornmalen, E.J.P.; Wat, J.; et al. Signaling of an Allosteric, Nanomolar Potent, Low Molecular Weight Agonist for the Follicle-Stimulating Hormone Receptor. Biochem. Pharmacol. 2013, 85, 1162–1170. [Google Scholar] [CrossRef]
- Sriraman, V.; Denis, D.; de Matos, D.; Yu, H.; Palmer, S.; Nataraja, S. Investigation of a Thiazolidinone Derivative as an Allosteric Modulator of Follicle Stimulating Hormone Receptor: Evidence for Its Ability to Support Follicular Development and Ovulation. Biochem. Pharmacol. 2014, 89, 266–275. [Google Scholar] [CrossRef]
- Yu, H.N.; Richardson, T.E.; Nataraja, S.; Fischer, D.J.; Sriraman, V.; Jiang, X.; Bharathi, P.; Foglesong, R.J.; Haxell, T.F.N.; Heasley, B.H.; et al. Discovery of Substituted Benzamides as Follicle Stimulating Hormone Receptor Allosteric Modulators. Bioorg. Med. Chem. Lett. 2014, 24, 2168–2172. [Google Scholar] [CrossRef]
- Nataraja, S.; Yu, H.; Guner, J.; Palmer, S. Discovery and Preclinical Development of Orally Active Small Molecules That Exhibit Highly Selective Follicle Stimulating Hormone Receptor Agonism. Front. Pharmacol. 2021, 11, 602593. [Google Scholar] [CrossRef]
- Danesi, R.; La Rocca, R.V.; Cooper, M.R.; Ricciardi, M.P.; Pellegrini, A.; Soldani, P.; Kragel, P.J.; Paparelli, A.; Del Tacca, M.; Myers, C.E. Clinical and Experimental Evidence of Inhibition of Testosterone Production by Suramin. J. Clin. Endocrinol. Metab. 1996, 81, 2238–2246. [Google Scholar] [CrossRef][Green Version]
- Yanofsky, S.D.; Shen, E.S.; Holden, F.; Whitehorn, E.; Aguilar, B.; Tate, E.; Holmes, C.P.; Scheuerman, R.; MacLean, D.; Wu, M.M.; et al. Allosteric Activation of the Follicle-Stimulating Hormone (FSH) Receptor by Selective, Nonpeptide Agonists. J. Biol. Chem. 2006, 281, 13226–13233. [Google Scholar] [CrossRef]
- Anderson, R.C.; Newton, C.L.; Millar, R.P. Small Molecule Follicle-Stimulating Hormone Receptor Agonists and Antagonists. Front. Endocrinol. 2019, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, J.; Jetter, J.; Kao, W.; Rogers, J.; Di, L.; Chi, J.; Peréz, M.C.; Chen, G.-C.; Shen, E.S. 5-Alkylated Thiazolidinones as Follicle-Stimulating Hormone (FSH) Receptor Agonists. Bioorg. Med. Chem. 2006, 14, 5729–5741. [Google Scholar] [CrossRef] [PubMed]
- Arey, B.J.; Yanofsky, S.D.; Claudia Pérez, M.; Holmes, C.P.; Wrobel, J.; Gopalsamy, A.; Stevis, P.E.; López, F.J.; Winneker, R.C. Differing Pharmacological Activities of Thiazolidinone Analogs at the FSH Receptor. Biochem. Biophys. Res. Commun. 2008, 368, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Sposini, S.; De Pascali, F.; Richardson, R.; Sayers, N.S.; Perrais, D.; Yu, H.N.; Palmer, S.; Nataraja, S.; Reiter, E.; Hanyaloglu, A.C. Pharmacological Programming of Endosomal Signaling Activated by Small Molecule Ligands of the Follicle Stimulating Hormone Receptor. Front. Pharmacol. 2020, 11, 593492. [Google Scholar] [CrossRef] [PubMed]
- Klein Herenbrink, C.; Sykes, D.A.; Donthamsetti, P.; Canals, M.; Coudrat, T.; Shonberg, J.; Scammells, P.J.; Capuano, B.; Sexton, P.M.; Charlton, S.J.; et al. The Role of Kinetic Context in Apparent Biased Agonism at GPCRs. Nat. Commun. 2016, 7, 10842. [Google Scholar] [CrossRef] [PubMed]
- Piketty, V.; Kara, E.; Guillou, F.; Reiter, E.; Crepieux, P. Follicle-Stimulating Hormone (FSH) Activates Extracellular Signal-Regulated Kinase Phosphorylation Independently of Beta-Arrestin- and Dynamin-Mediated FSH Receptor Internalization. Reprod. Biol. Endocrinol. RBE 2006, 4, 33. [Google Scholar] [CrossRef]
- Sposini, S.; Jean-Alphonse, F.G.; Ayoub, M.A.; Oqua, A.; West, C.; Lavery, S.; Brosens, J.J.; Reiter, E.; Hanyaloglu, A.C. Integration of GPCR Signaling and Sorting from Very Early Endosomes via Opposing APPL1 Mechanisms. Cell Rep. 2017, 21, 2855–2867. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Wang, J.; Plouffe, B.; Smith, J.S.; Yamani, L.; Kaur, S.; Jean-Charles, P.-Y.; Gauthier, C.; Lee, M.-H.; Pani, B.; et al. Manifold Roles of β-Arrestins in GPCR Signaling Elucidated with SiRNA and CRISPR/Cas9. Sci. Signal. 2018, 11, eaat7650. [Google Scholar] [CrossRef]
- Riccetti, L.; De Pascali, F.; Gilioli, L.; Santi, D.; Brigante, G.; Simoni, M.; Casarini, L. Genetics of Gonadotropins and Their Receptors as Markers of Ovarian Reserve and Response in Controlled Ovarian Stimulation. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 44, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, M.G.F.; Kramer, H.; el Galta, R.; van Beerendonk, G.; Hanssen, R.; Abd-Elaziz, K.; Klipping, C.; Duijkers, I.; Stoch, S.A. Oral Follicle-Stimulating Hormone Agonist Tested in Healthy Young Women of Reproductive Age Failed to Demonstrate Effect on Follicular Development but Affected Thyroid Function. Fertil. Steril. 2016, 105, 1056–1062.e4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wan, Q.; Okashah, N.; Inoue, A.; Nehmé, R.; Carpenter, B.; Tate, C.G.; Lambert, N.A. Mini G Protein Probes for Active G Protein-Coupled Receptors (GPCRs) in Live Cells. J. Biol. Chem. 2018, 293, 7466–7473. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, M.A.; Landomiel, F.; Gallay, N.; Jégot, G.; Poupon, A.; Crépieux, P.; Reiter, E. Assessing Gonadotropin Receptor Function by Resonance Energy Transfer-Based Assays. Front. Endocrinol. 2015, 6, 130. [Google Scholar] [CrossRef]
- Daaka, Y.; Luttrell, L.M.; Lefkowitz, R.J. Switching of the Coupling of the Beta2-Adrenergic Receptor to Different G Proteins by Protein Kinase A. Nature 1997, 390, 88–91. [Google Scholar] [CrossRef]
- Zamah, A.M.; Delahunty, M.; Luttrell, L.M.; Lefkowitz, R.J. Protein Kinase A-Mediated Phosphorylation of the Beta 2-Adrenergic Receptor Regulates Its Coupling to Gs and Gi. Demonstration in a Reconstituted System. J. Biol. Chem. 2002, 277, 31249–31256. [Google Scholar] [CrossRef]
- Rajagopal, S.; Ahn, S.; Rominger, D.H.; Gowen-MacDonald, W.; Lam, C.M.; Dewire, S.M.; Violin, J.D.; Lefkowitz, R.J. Quantifying Ligand Bias at Seven-Transmembrane Receptors. Mol. Pharmacol. 2011, 80, 367–377. [Google Scholar] [CrossRef]
- Kenakin, T.; Watson, C.; Muniz-Medina, V.; Christopoulos, A.; Novick, S. A Simple Method for Quantifying Functional Selectivity and Agonist Bias. ACS Chem. Neurosci. 2012, 3, 193–203. [Google Scholar] [CrossRef]
- Stahl, E.L.; Zhou, L.; Ehlert, F.J.; Bohn, L.M. A Novel Method for Analyzing Extremely Biased Agonism at G Protein-Coupled Receptors. Mol. Pharmacol. 2015, 87, 866–877. [Google Scholar] [CrossRef] [PubMed]
- O’Hayre, M.; Eichel, K.; Avino, S.; Zhao, X.; Steffen, D.J.; Feng, X.; Kawakami, K.; Aoki, J.; Messer, K.; Sunahara, R.; et al. Genetic Evidence That β-Arrestins Are Dispensable for the Initiation of Β2-Adrenergic Receptor Signaling to ERK. Sci. Signal. 2017, 10, eaal3395. [Google Scholar] [CrossRef] [PubMed]
- Stallaert, W.; van der Westhuizen, E.T.; Schönegge, A.-M.; Plouffe, B.; Hogue, M.; Lukashova, V.; Inoue, A.; Ishida, S.; Aoki, J.; Le Gouill, C.; et al. Purinergic Receptor Transactivation by the Β2-Adrenergic Receptor Increases Intracellular Ca2+ in Nonexcitable Cells. Mol. Pharmacol. 2017, 91, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Grundmann, M.; Merten, N.; Malfacini, D.; Inoue, A.; Preis, P.; Simon, K.; Rüttiger, N.; Ziegler, N.; Benkel, T.; Schmitt, N.K.; et al. Lack of Beta-Arrestin Signaling in the Absence of Active G Proteins. Nat. Commun. 2018, 9, 341. [Google Scholar] [CrossRef]
- Jiang, L.I.; Collins, J.; Davis, R.; Lin, K.-M.; DeCamp, D.; Roach, T.; Hsueh, R.; Rebres, R.A.; Ross, E.M.; Taussig, R.; et al. Use of a CAMP BRET Sensor to Characterize a Novel Regulation of CAMP by the Sphingosine 1-Phosphate/G13 Pathway. J. Biol. Chem. 2007, 282, 10576–10584. [Google Scholar] [CrossRef]
- Troispoux, C.; Guillou, F.; Elalouf, J.M.; Firsov, D.; Iacovelli, L.; De Blasi, A.; Combarnous, Y.; Reiter, E. Involvement of G Protein-Coupled Receptor Kinases and Arrestins in Desensitization to Follicle-Stimulating Hormone Action. Mol. Endocrinol. 1999, 13, 1599–1614. [Google Scholar] [CrossRef]
- Hoare, S.R.J.; Pierre, N.; Moya, A.G.; Larson, B. Kinetic Operational Models of Agonism for G-Protein-Coupled Receptors. J. Theor. Biol. 2018, 446, 168–204. [Google Scholar] [CrossRef] [PubMed]
- van der Westhuizen, E.T.; Breton, B.; Christopoulos, A.; Bouvier, M. Quantification of Ligand Bias for Clinically Relevant Β2-Adrenergic Receptor Ligands: Implications for Drug Taxonomy. Mol. Pharmacol. 2014, 85, 492–509. [Google Scholar] [CrossRef]
- Raue, A.; Steiert, B.; Schelker, M.; Kreutz, C.; Maiwald, T.; Hass, H.; Vanlier, J.; Tönsing, C.; Adlung, L.; Engesser, R.; et al. Data2Dynamics: A Modeling Environment Tailored to Parameter Estimation in Dynamical Systems. Bioinforma. Oxf. Engl. 2015, 31, 3558–3560. [Google Scholar] [CrossRef] [PubMed]
- Raue, A.; Karlsson, J.; Saccomani, M.P.; Jirstrand, M.; Timmer, J. Comparison of Approaches for Parameter Identifiability Analysis of Biological Systems. Bioinforma. Oxf. Engl. 2014, 30, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Practical Guide to Cluster Analysis in R: Unsupervised Machine Learning, 1st ed.; Multivariate analysis; STHDA: France, 2017; ISBN 978-1-5424-6270-9. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Pascali, F.; Ayoub, M.A.; Benevelli, R.; Sposini, S.; Lehoux, J.; Gallay, N.; Raynaud, P.; Landomiel, F.; Jean-Alphonse, F.; Gauthier, C.; et al. Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor. Int. J. Mol. Sci. 2021, 22, 9850. https://doi.org/10.3390/ijms22189850
De Pascali F, Ayoub MA, Benevelli R, Sposini S, Lehoux J, Gallay N, Raynaud P, Landomiel F, Jean-Alphonse F, Gauthier C, et al. Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor. International Journal of Molecular Sciences. 2021; 22(18):9850. https://doi.org/10.3390/ijms22189850
Chicago/Turabian StyleDe Pascali, Francesco, Mohammed Akli Ayoub, Riccardo Benevelli, Silvia Sposini, Jordan Lehoux, Nathalie Gallay, Pauline Raynaud, Flavie Landomiel, Frédéric Jean-Alphonse, Christophe Gauthier, and et al. 2021. "Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor" International Journal of Molecular Sciences 22, no. 18: 9850. https://doi.org/10.3390/ijms22189850
APA StyleDe Pascali, F., Ayoub, M. A., Benevelli, R., Sposini, S., Lehoux, J., Gallay, N., Raynaud, P., Landomiel, F., Jean-Alphonse, F., Gauthier, C., Pellissier, L. P., Crépieux, P., Poupon, A., Inoue, A., Joubert, N., Viaud-Massuard, M.-C., Casarini, L., Simoni, M., Hanyaloglu, A. C., ... Reiter, E. (2021). Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor. International Journal of Molecular Sciences, 22(18), 9850. https://doi.org/10.3390/ijms22189850







