Bitter Taste Receptor T2R14 Modulates Gram-Positive Bacterial Internalization and Survival in Gingival Epithelial Cells
Abstract
:1. Introduction
2. Results
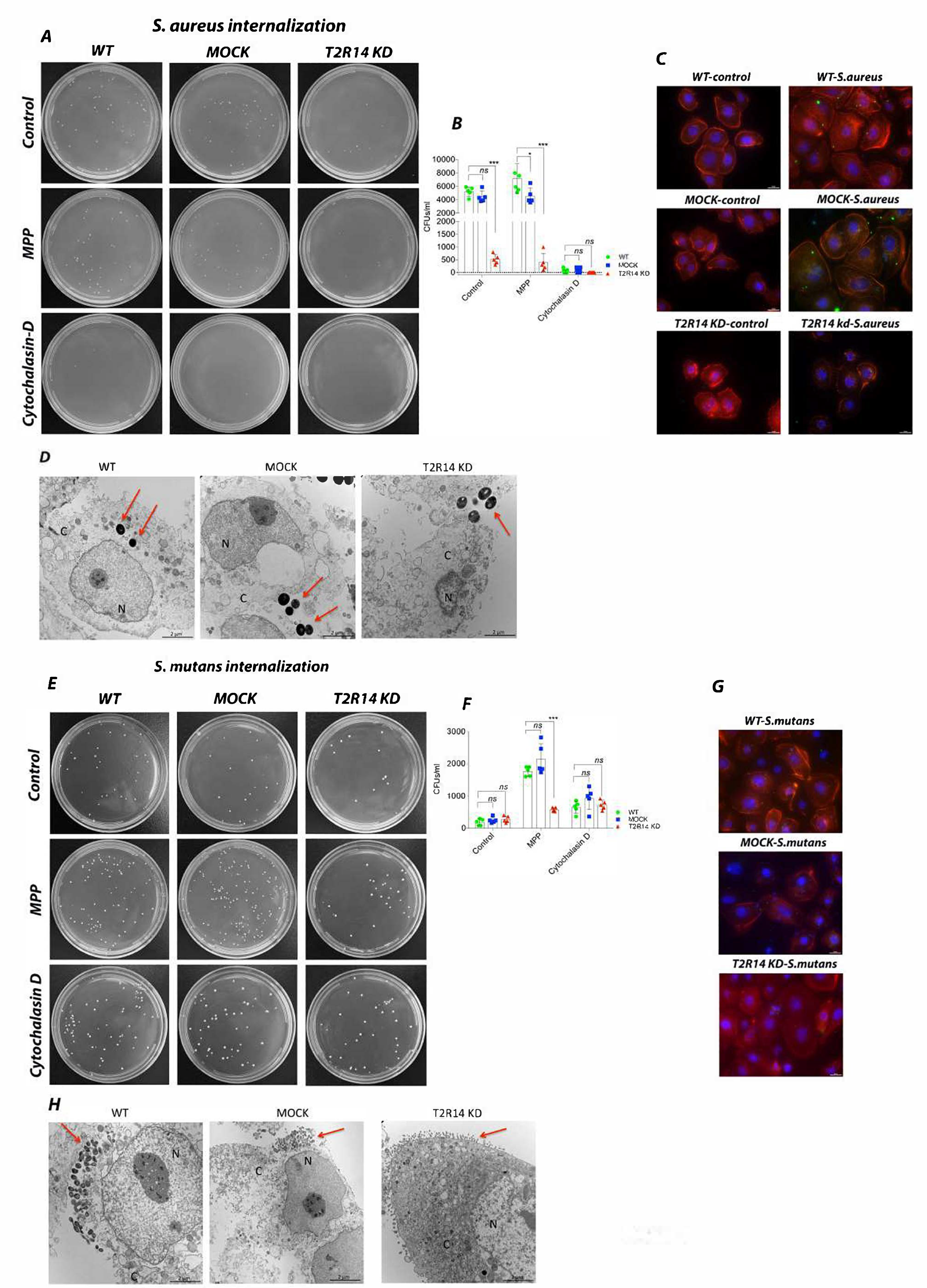
2.1. T2R14 Mediates Internalization of S. aureus and S. mutans in GECs
2.2. Knockdown of T2R14 Decreases PAK1 Associated Actin and F-Actin but Not GTP-Rac1 in GECs
2.3. S. aureus–Induced hBD-2 Secretion Is T2R14 Dependent
2.4. Gram-Positive Bacteria Do Not Induce Nitrite/Nitrate Secretion in GECs
2.5. IL-8/CXCL-8 Secretion in GECs Is T2R14-Dependent
2.6. Activation of T2R14 in GECs by CSP-1 and DPH has a Growth Inhibitory Effect on S. aureus but Not S. mutans
3. Discussion
4. Materials and Methods
4.1. Reagents Used in the Study
4.2. Cell Line Used in the Study
4.3. Bacterial Strains Used in the Study
4.4. Bacterial Internalization Assay
4.5. Immunofluorescence Microscopy of Internalized Bacteria in Host Cells
4.6. Transmission Electron Microscopy Imaging of GECs
4.7. GTP-Rac1 Pull-Down Assay
4.8. Isolation and Quantification of F/G-Actins in GECs
4.9. Bacterial Survival Assay
4.10. Human β-Defensin 2 (hBD-2) ELISA
4.11. Measurement of Nitrate/Nitrite Secretion
4.12. Measurement of IL-8/CXCL8 Secretion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ooi, E.H.; Wormald, P.-J.; Tan, L.W. Innate Immunity in the Paranasal Sinuses: A Review of Nasal Host Defenses. Am. J. Rhinol. 2008, 22, 13–19. [Google Scholar] [CrossRef]
- Medapati, M.R.; Singh, N.; Bhagirath, A.Y.; Duan, K.; Triggs-Raine, B.; Batista, E.L.; Chelikani, P. Bitter taste receptor T2R14 detects quorum sensing molecules from cariogenic Streptococcus mutans and mediates innate immune responses in gingival epithelial cells. FASEB J. 2021, 35, e21375. [Google Scholar] [CrossRef]
- Jaggupilli, A.; Singh, N.; De Jesus, V.C.; Duan, K.; Chelikani, P. Characterization of the Binding Sites for Bacterial Acyl Homoserine Lactones (AHLs) on Human Bitter Taste Receptors (T2Rs). ACS Infect. Dis. 2018, 4, 1146–1156. [Google Scholar] [CrossRef]
- Freund, J.; Mansfield, C.J.; Doghramji, L.J.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Reed, D.R.; Jiang, P.; Lee, R.J. Activation of airway epithelial bitter taste receptors by Pseudomonas aeruginosa quinolones modulates calcium, cyclic-AMP, and nitric oxide signaling. J. Biol. Chem. 2018, 293, 9824–9840. [Google Scholar] [CrossRef] [Green Version]
- Lindemann, B. Taste reception. Physiol. Rev. 1996, 76, 719–766. [Google Scholar] [CrossRef]
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.; Zuker, C.S. A Novel Family of Mammalian Taste Receptors. Cell 2000, 100, 693–702. [Google Scholar] [CrossRef] [Green Version]
- Jaggupilli, A.; Singh, N.; Upadhyaya, J.; Arakawa, M.; Dakshinamurti, S.; Duan, K.; Chelikani, P.; Sikarwar, A.S.; Bhullar, R.P. Analysis of the expression of human bitter taste receptors in extraoral tissues. Mol. Cell. Biochem. 2017, 426, 137–147. [Google Scholar] [CrossRef]
- Shaik, F.A.; Singh, N.; Arakawa, M.; Duan, K.; Bhullar, R.P.; Chelikani, P. Bitter taste receptors: Extraoral roles in pathophysiology. Int. J. Biochem. Cell Biol. 2016, 77, 197–204. [Google Scholar] [CrossRef]
- Carey, R.M.; Workman, A.D.; Yan, C.; Chen, B.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Lee, R.; Cohen, N.A. Sinonasal T2R-Mediated Nitric Oxide Production in Response to Bacillus Cereus. Am. J. Rhinol. Allergy 2017, 31, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.; Xiong, G.; Kofonow, J.M.; Chen, B.; Lysenko, A.; Jiang, P.; Abraham, V.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Investig. 2012, 122, 4145–4159. [Google Scholar] [CrossRef] [Green Version]
- Gopallawa, I.; Freund, J.; Lee, R.J. Bitter taste receptors stimulate phagocytosis in human macrophages through calcium, nitric oxide, and cyclic-GMP signaling. Cell. Mol. Life Sci. 2021, 78, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.S.; Ben-Shahar, Y.; Moninger, T.O.; Kline, J.N.; Welsh, M.J. Motile Cilia of Human Airway Epithelia Are Chemosensory. Science 2009, 325, 1131–1134. [Google Scholar] [CrossRef] [Green Version]
- Howitt, M.R.; Lavoie, S.; Michaud, M.; Blum, A.M.; Tran, S.V.; Weinstock, J.V.; Gallini, C.A.; Redding, K.; Margolskee, R.F.; Osborne, L.C.; et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016, 351, 1329–1333. [Google Scholar] [CrossRef] [Green Version]
- Von Moltke, J.; Ji, M.; Liang, H.E.; Locksley, R.M. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 2016, 529, 221–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malki, A.; Fiedler, J.; Fricke, K.; Ballweg, I.; Pfaffl, M.; Krautwurst, D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J. Leukoc. Biol. 2015, 97, 533–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaida, M.M.; Dapunt, U.; Hänsch, G.M. Sensing developing biofilms: The bitter receptor T2R38 on myeloid cells. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekoff, M.; Choi, J.H.; James, A.; Dahlén, B.; Nilsson, G.; Dahlén, S.E. Bitter taste receptor (TAS2R) agonists inhibit IgE-dependent mast cell activation. J. Allergy Clin. Immunol. 2014, 134, 475–478. [Google Scholar] [CrossRef]
- De Jesus, V.C.; Khan, M.W.; Mittermuller, B.A.; Duan, K.; Hu, P.; Schroth, R.J.; Chelikani, P. Characterization of Supragingival Plaque and Oral Swab Microbiomes in Children With Severe Early Childhood Caries. Front. Microbiol. 2021, 12, 683–685. [Google Scholar] [CrossRef]
- Fowler, T.; Johansson, S.; Wary, K.K.; Höök, M. Src kinase has a central role in in vitro cellular internalization of Staphylococcus aureus. Cell. Microbiol. 2003, 5, 417–426. [Google Scholar] [CrossRef]
- Agerer, F.; Lux, S.; Michel, A.; Rohde, M.; Ohlsen, K.; Hauck, C.R. Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalisation. J. Cell Sci. 2005, 118, 2189–2200. [Google Scholar] [CrossRef] [Green Version]
- Selbach, M.; Backert, S. Cortactin: An Achilles’ heel of the actin cytoskeleton targeted by pathogens. Trends Microbiol. 2005, 13, 181–189. [Google Scholar] [CrossRef]
- Lehrer, R.I. Primate defensins. Nat. Rev. Microbiol. 2004, 2, 727–738. [Google Scholar] [CrossRef]
- Carey, R.M.; Chen, B.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Lee, R.J.; Cohen, N.A. Human upper airway epithelium produces nitric oxide in response to Staphylococcus epidermidis. Int. Forum Allergy Rhinol. 2016, 6, 1238–1244. [Google Scholar] [CrossRef] [Green Version]
- Hariri, B.M.; McMahon, D.B.; Chen, B.; Freund, J.; Mansfield, C.J.; Doghramji, L.J.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Reed, D.R.; et al. Flavones modulate respiratory epithelial innate immunity: Anti-inflammatory effects and activation of the T2R14 receptor. J. Biol. Chem. 2017, 292, 8484–8497. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, C.; Jaggupilli, A.; Chelikani, P.; Bhullar, R.P. Regulation of Rac1 GTPase activity by quinine through G-protein and bitter taste receptor T2R4. Mol. Cell. Biochem. 2017, 426, 129–136. [Google Scholar] [CrossRef]
- Vidal, C.; Geny, B.; Melle, J.; Jandrot-Perrus, M.; Fontenay, M. Cdc42/Rac1-dependent activation of the p21-activated kinase (PAK) regulates human platelet lamellipodia spreading: Implication of the cortical-actin binding protein cortactin. Blood 2002, 100, 4462–4469. [Google Scholar] [CrossRef] [Green Version]
- Grassart, A.; Meas-Yedid, V.; Dufour, A.; Olivo-Marin, J.-C.; Dautry-Varsat, A.; Sauvonnet, N. Pak1 Phosphorylation Enhances Cortactin-N-WASP Interaction in Clathrin-Caveolin-Independent Endocytosis. Traffic 2010, 11, 1079–1091. [Google Scholar] [CrossRef]
- Zheng, X.; Tizzano, M.; Redding, K.; He, J.; Peng, X.; Jiang, P.; Xu, X.; Zhou, X.; Margolskee, R.F. Gingival solitary chemosensory cells are immune sentinels for periodontitis. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, S.; Coldwell, S.; Drury, J.L.; Arroyo, F.; Phi, T.; Saadat, S.; Kwong, D.; Chung, W.O. Genotype-specific regulation of oral innate immunity by T2R38 taste receptor. Mol. Immunol. 2015, 68, 663–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, W.O.; Dale, B.A. Innate Immune Response of Oral and Foreskin Keratinocytes: Utilization of Different Signaling Pathways by Various Bacterial Species. Infect. Immun. 2004, 72, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Carey, R.M.; Ba, A.D.W.; Hatten, K.M.; Siebert, A.P.; Brooks, S.G.; Chen, B.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Lee, R.; et al. Denatonium-induced sinonasal bacterial killing may play a role in chronic rhinosinusitis outcomes. Int. Forum Allergy Rhinol. 2017, 7, 699–704. [Google Scholar] [CrossRef]
- Lee, R.; Kofonow, J.M.; Rosen, P.L.; Siebert, A.P.; Chen, B.; Doghramji, L.; Xiong, G.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; et al. Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J. Clin. Investig. 2014, 124, 1393–1405. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.J.; Ahn, S.J.; Wen, Z.T.; Brady, L.J.; Burne, R.A. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 2008, 76, 4259–4268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Kudo, Y.; Baker, J.L.; LaBonte, S.; Jordan, P.A.; McKinnie, S.M.K.; Guo, J.; Huan, T.; Moore, B.S.; Edlund, A. Cariogenic Streptococcus mutans Produces Tetramic Acid Strain-Specific Antibiotics That Impair Commensal Colonization. ACS Infect. Dis. 2020, 6, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A.; Hahn, W.C.; Ino, Y.; Ronfard, V.; Wu, J.Y.; Weinberg, R.A.; Louis, D.N.; Li, F.P.; Rheinwald, J.G. Human Keratinocytes That Express hTERT and Also Bypass a p16 INK4a -Enforced Mechanism That Limits Life Span Become Immortal yet Retain Normal Growth and Differentiation Characteristics. Mol. Cell. Biol. 2000, 20, 1436–1447. [Google Scholar] [CrossRef] [Green Version]
- McMahon, L.; Schwartz, K.; Yilmaz, O.; Brown, E.; Ryan, L.K.; Diamond, G. Vitamin D-Mediated Induction of Innate Immunity in Gingival Epithelial Cells. Infect. Immun. 2011, 79, 2250–2256. [Google Scholar] [CrossRef] [Green Version]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.-Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Al Kindi, A.; Alkahtani, A.M.; Nalubega, M.; El-Chami, C.; O’Neill, C.; Arkwright, P.D.; Pennock, J.L. Staphylococcus aureus Internalized by Skin Keratinocytes Evade Antibiotic Killing. Front. Microbiol. 2019, 10, 2242. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Chevrier, J.; Dubus, M.; Varin-Simon, J.; Sergheraert, J.; Gangloff, S.C.; Reffuveille, F.; Mauprivez, C.; Kerdjoudj, H. Infection of Human Dental Pulp Stromal Cells by Streptococcus mutans: Shedding Light on Bacteria Pathogenicity and Pulp Inflammation. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Wunsch, C.M.; Lewis, J.P. Porphyromonas gingivalis as a Model Organism for Assessing Interaction of Anaerobic Bacteria with Host Cells. J. Vis. Exp. 2015, 53408. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medapati, M.R.; Bhagirath, A.Y.; Singh, N.; Schroth, R.J.; Bhullar, R.P.; Duan, K.; Chelikani, P. Bitter Taste Receptor T2R14 Modulates Gram-Positive Bacterial Internalization and Survival in Gingival Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 9920. https://doi.org/10.3390/ijms22189920
Medapati MR, Bhagirath AY, Singh N, Schroth RJ, Bhullar RP, Duan K, Chelikani P. Bitter Taste Receptor T2R14 Modulates Gram-Positive Bacterial Internalization and Survival in Gingival Epithelial Cells. International Journal of Molecular Sciences. 2021; 22(18):9920. https://doi.org/10.3390/ijms22189920
Chicago/Turabian StyleMedapati, Manoj Reddy, Anjali Yadav Bhagirath, Nisha Singh, Robert J. Schroth, Rajinder P. Bhullar, Kangmin Duan, and Prashen Chelikani. 2021. "Bitter Taste Receptor T2R14 Modulates Gram-Positive Bacterial Internalization and Survival in Gingival Epithelial Cells" International Journal of Molecular Sciences 22, no. 18: 9920. https://doi.org/10.3390/ijms22189920





