Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of JAZ Genes in Tomato
2.2. Phylogenetic Relationships and Gene Structure Analysis of SlJAZs
2.3. Detection of Cis-Acting Elements in the Promoter Regions of SlJAZ Genes
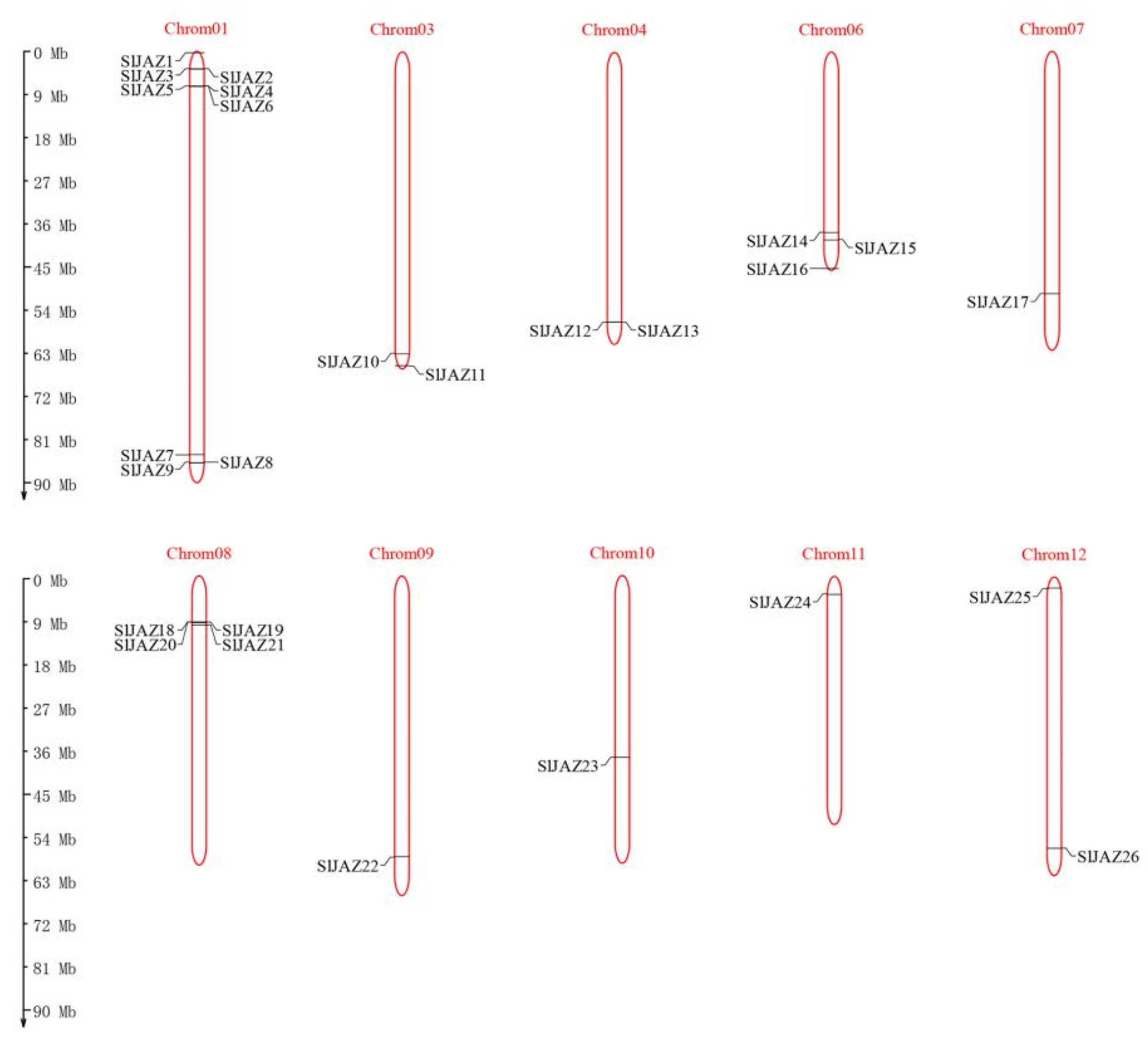
2.4. Gene Duplication and Synteny Analysis of SlJAZ Genes
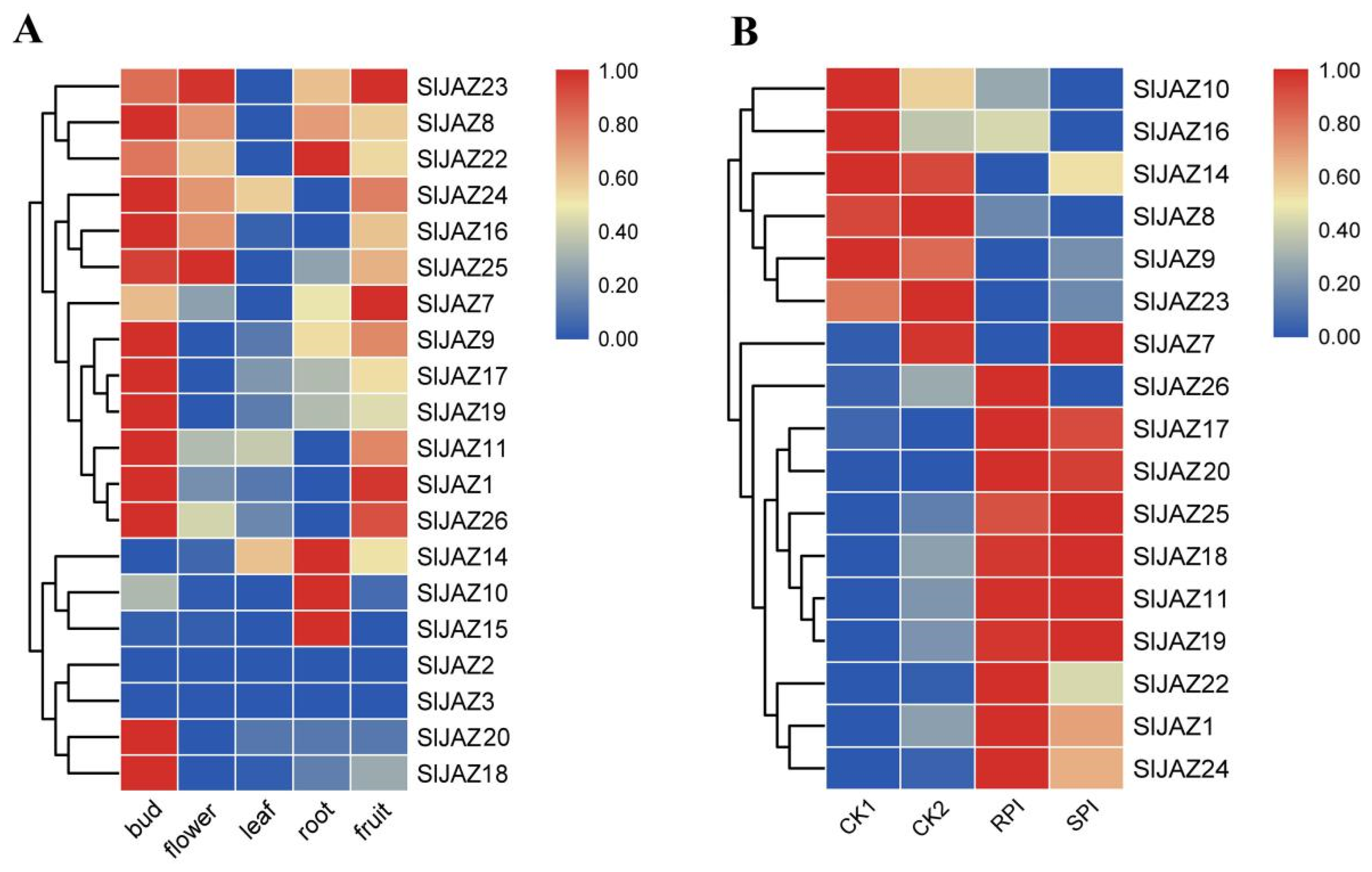
2.5. Spatiotemporal Expression Characteristics of SlJAZ Genes in Tomato
2.6. Expression Patterns of SlJAZ Genes According to RNA-seq after Inoculation with S. lycopersici
2.7. Expression Profiles of SlJAZ Genes as Assessed via qRT-PCR
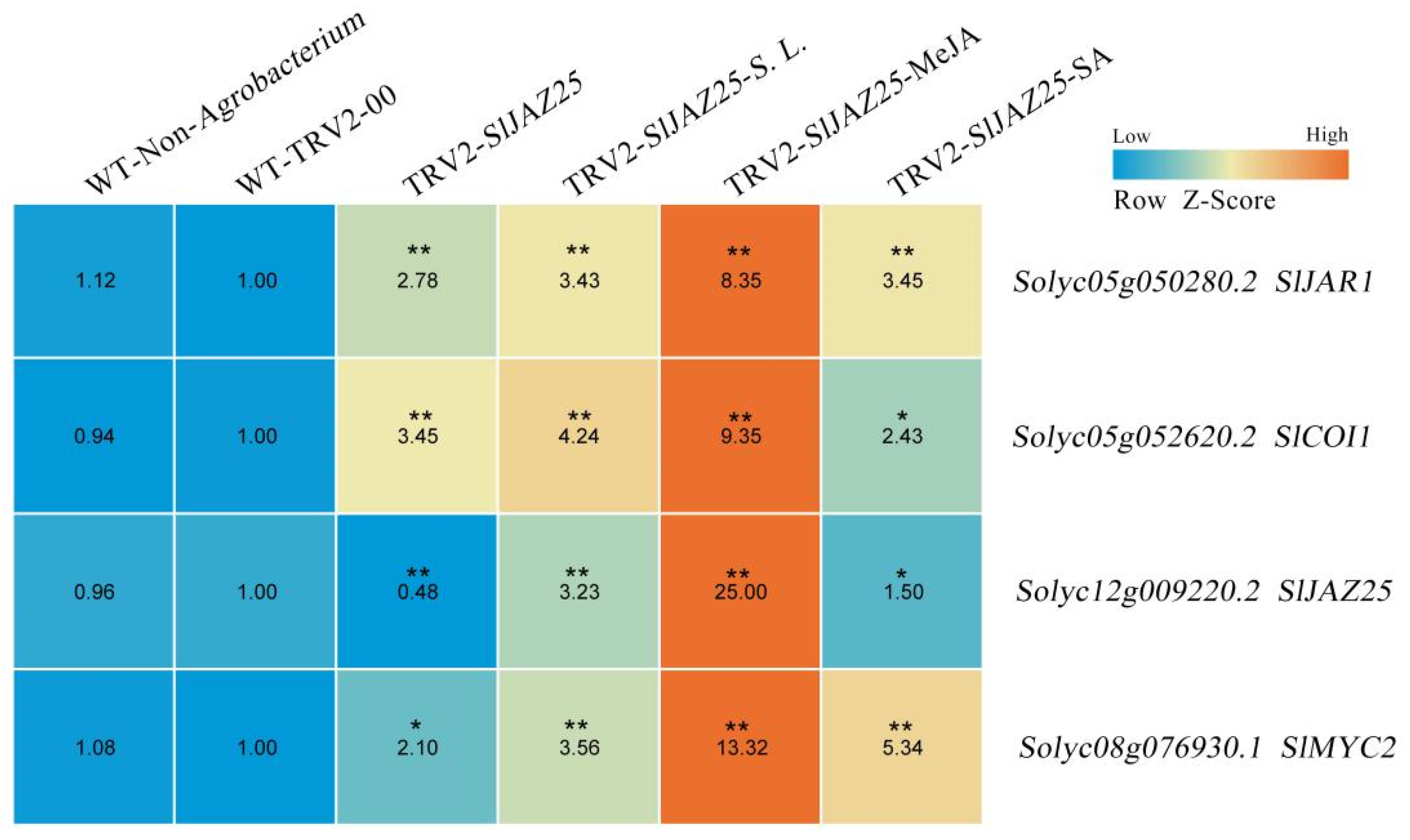
2.8. Effects of SlJAZ25 Silencing on the S. lycopersici Defense Response
2.9. Analysis of the Expression Patterns of Pathway Genes Related to SlJAZ25
2.10. Analysis of the Subcellular Localization of SlJAZ25
2.11. Analysis of the SlJAZ Gene Co-Expression Network and Yeast One-Hybrid System
3. Discussion
3.1. Effect of the JA Pathway on Plant Disease Resistance
3.2. Distribution of the JAZ Gene Family in Plants
3.3. Classification and Structure of JAZ Genes in Tomato
3.4. The Promoters of SlJAZ Genes Contain a Large Number of Cis-Acting Elements Related to Disease Resistance
3.5. Intraspecific and Interspecific Relationship of SlJAZ Genes
3.6. Transcriptome Combined with qRT-PCR Techniques to Reveal the Expression Profile of SlJAZ Genes
3.7. Resistance of the SlJAZ25 Gene to Gray Leaf Spots in Tomato and Its Hypothetical Pattern
3.8. Synergism between JA and Other Hormones
4. Materials and Methods
4.1. Plant Growth and Treatments
4.2. Identification of JAZ Family Members in Tomato
4.3. Chromosomal Mapping, Phylogenetic Relationship Analysis, and Gene Sequence Analysis of JAZ Gene Family Members in Tomato
4.4. Analysis of Cis-Acting Elements in the SlJAZ Gene Promoter
4.5. Gene Duplication and Homeology Analysis of SlJAZ Genes
4.6. Analysis of SlJAZ Gene Expression Patterns via Transcriptomic Data
4.7. VIGS Vector Construction and Agroinfiltration
4.8. Determination of Gene Silencing Efficiency and Phenotypic Observations
4.9. Subcellular Localization of SlJAZ25
4.10. Analysis of the Expression Network of SlJAZs in Tomato
4.11. Yeast One-Hybrid Assays
4.12. Total RNA Extraction, cDNA Synthesis and qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jander, G.; Clay, N. New Synthesis–Plant Defense Signaling: New Opportunities for Studying Chemical Diversity. J. Chem. Ecol. 2011, 37, 429. [Google Scholar] [CrossRef] [Green Version]
- Kud, J.; Wang, W.; Gross, R.; Fan, Y.; Huang, L.; Yuan, Y.; Gray, A.; Duarte, A.; Kuhl, J.C.; Caplan, A.; et al. The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 2019, 15, e1007720. [Google Scholar] [CrossRef] [Green Version]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [Green Version]
- Ohta, T. Evolution of gene families. Gene 2000, 259, 45–52. [Google Scholar] [CrossRef]
- Galiano, L.M.J.; Hernández, G.A.I.; Salvador, C.O.; Rausell, C.; Real, M.D.; Escamilla, M.; Camañes, G.; Agustín, G.P.; Bosch, G.C.; Robles, G.I. Epigenetic regulation of the expression of WRKY75 transcription factor in response to biotic and abiotic stresses in Solanaceae plants. Plant Cell Rep. 2018, 37, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Benoit, M.; Drost, H.G.; Catoni, M.; Gouil, Q.; Gomollon, L.S.; Baulcombe, D.; Paszkowski, J. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PLoS Genet. 2019, 15, e1008370. [Google Scholar] [CrossRef] [Green Version]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; Oliveira, F.D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Demianski, A.J.; Chung, K.M.; Kunkel, B.N. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2012, 13, 46–57. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, D.; Wang, Y.; Xie, D. Jasmonate action in plant defense against insects. J. Exp. Bot. 2019, 70, 3391–3400. [Google Scholar] [CrossRef]
- Huang, R.; Li, Y.; Tang, G.; Hui, S.; Yang, Z.; Zhao, J.; Liu, H.; Cao, J.; Yuan, M. Dynamic phytohormone profiling of rice upon rice black-streaked dwarf virus invasion. J. Plant Physiol. 2018, 228, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Liechti, R.; Farmer, E.E. Arabidopsis jasmonate signaling pathway. Sci. Signal. 2010, 3, cm4. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G. Jasmonates: Biosynthesis, perception and signal transduction. Essays Biochem. 2020, 64, 501–512. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Zhang, L.; Wei, X.; Zhou, Y.; Dai, Y.; Zhu, Z. OsJAZ13 Negatively Regulates Jasmonate Signaling and Activates Hypersensitive Cell Death Response in Rice. Int. J. Mol. Sci. 2020, 21, 4379. [Google Scholar] [CrossRef]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genomics 2021, 22, 256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, W.; Niu, D.; Wang, Z.; Yan, X.; Yang, X.; Yang, Y.; Cui, H. Tobacco transcription repressors NtJAZ: Potential involvement in abiotic stress response and glandular trichome induction. Plant Physiol. Biochem. 2019, 141, 388–397. [Google Scholar] [CrossRef]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef]
- Sun, J.Q.; Jiang, H.L.; Li, C.Y. Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 2011, 4, 607–615. [Google Scholar] [CrossRef]
- Chini, A.; Romdhane, B.W.; Hassairi, A.; Soud, A.M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef] [PubMed]
- Ishiga, Y.; Ishiga, T.; Uppalapati, S.R.; Mysore, K.S. Jasmonate ZIM-domain (JAZ) protein regulates host and nonhost pathogen-induced cell death in tomato and Nicotiana benthamiana. PLoS ONE 2013, 8, e75728. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods 2020, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, T.; Liu, M.; Nie, Z.; Yang, H.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Virus-induced gene silencing of SlPKY1 attenuates defense responses against gray leaf spot in tomato. Sci. Hortic. 2020, 264, 109149. [Google Scholar] [CrossRef]
- Medina, R.; Franco, M.E.E.; Cabral, L.D.; Bahima, J.V.; Patriarca, A.; Balatti, P.A.; Saparrat, M.C.N. The secondary metabolites profile of Stemphylium lycopersici, the causal agent of tomato grey leaf spot, is complex and includes host and non-host specific toxins. Australas. Plant Pathol. 2021, 50, 105–115. [Google Scholar] [CrossRef]
- Saito, R.; Hayashi, K.; Nomoto, H.; Nakayama, M.; Takaoka, Y.; Saito, H.; Yamagami, S.; Muto, T.; Ueda, M. Extended JAZ degron sequence for plant hormone binding in jasmonate co-receptor of tomato SlCOI1-SlJAZ. Sci. Rep. 2021, 11, 13612. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Valenzuela-Riffo, F.; Figueroa, C.R. Evolutionary Analysis of JAZ Proteins in Plants: An Approach in Search of the Ancestral Sequence. Int. J. Mol. Sci. 2019, 20, 5060. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.Y.; Sunter, G. Interaction between the transcription factor AtTIFY4B and begomovirus AL2 protein impacts pathogenicity. Plant Mol. Biol. 2014, 86, 185–200. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, C.; Yang, L.; Zhang, Y.; Wang, Y.; Fang, Z.; Lv, H. Genome-Wide Identification, Expression Profile of the TIFY Gene Family in Brassica oleracea var. capitata, and Their Divergent Response to Various Pathogen Infections and Phytohormone Treatments. Genes 2020, 11, 127. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Wang, G.; Zhang, X.; Zhang, X.; Qiao, P.; Long, L.; Yuan, Y.; Cai, Y. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, J.; Grady, K.o.; Chen, S.; Gurley, W. The C-terminal WD40 repeats on the TOPLESS co-repressor function as a protein-protein interaction surface. Plant Mol. Biol. 2019, 100, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.Y.; Zhang, J.; Jiang, B.; Chan, C.; Yu, J.H.; Lu, Y.P.; Chung, K.; Zimmerli, L. NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. J. Exp. Bot. 2019, 70, 1033–1047. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Zorrilla, F.J.M.; Casado, G.G.; Zamarreño, A.M.; Mina, G.J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Zabala, M.T.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 2016, 209, 1120–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thatcher, L.F.; Cevik, V.; Grant, M.; Zhai, B.; Jones, J.D.; Manners, J.M.; Kazan, K. Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J. Exp. Bot. 2016, 67, 2367–2386. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, J.; Ji, J.; Li, P.; Yu, L.; Allah, E.F.A.; Luo, Y.; Hu, L.; Hu, X. High Temperature Induces Expression of Tobacco Transcription Factor NtMYC2a to Regulate Nicotine and JA Biosynthesis. Front. Physiol. 2016, 7, 465. [Google Scholar] [CrossRef] [Green Version]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Zhao, T.; Jiang, J.; Wang, S.; Wang, A.; Li, J.; Xu, X. Mapping and screening of the tomato Stemphylium lycopersici resistance gene, Sm, based on bulked segregant analysis in combination with genome resequencing. BMC Plant Biol. 2017, 17, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Jiang, J.; Yang, H.; Zhao, T.; Xu, X.; Li, J. Virus-induced gene silencing (VIGS) of the NBS-LRR gene SLNLC1 compromises Sm-mediated disease resistance to Stemphylium lycopersici in tomato. Biochem. Biophys. Res. Commun. 2018, 503, 1524–1529. [Google Scholar] [CrossRef]
- Amri, K.A.; Sadi, A.M.A.; Shihi, A.A.; Nasehi, A.; Mahmooli, A.I.; Deadman, M.L. Population structure of Stemphylium lycopersici associated with leaf spot of tomato in a single field. Springerplus 2016, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Zhu, G.; Huang, Z.; Wang, X.; Guo, Y.; Li, B.; Du, Y.; Yang, W.; Gao, J. Fine mapping and molecular marker development of the Sm gene conferring resistance to gray leaf spot (Stemphylium spp.) in tomato. Appl. Genet. 2019, 132, 871–882. [Google Scholar] [CrossRef]
- Yang, Y.X.; Ahammed, G.J.; Wu, C.; Fan, S.Y.; Zhou, Y.H. Crosstalk among Jasmonate, Salicylate and Ethylene Signaling Pathways in Plant Disease and Immune Responses. Curr. Protein Pept. Sci. 2015, 16, 450–461. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef]
- Miller, R.N.G.; Alves, G.S.C.; Sluys, M.A.V. Plant immunity: Unravelling the complexity of plant responses to biotic stresses. Ann. Bot. 2017, 119, 681–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blázquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef] [Green Version]
- Seilaniantz, A.R.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.; Does, D.V.; Zamioudis, C.; Reyes, A.L.; Wees, S.C.M.V. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Xia, X.C.; Han, L.H.; Ni, P.; Yan, J.Q.; Tao, M.; Huang, G.Q.; Li, X.B. Genome-wide identification and characterization of JAZ gene family in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Souza, S.J.; Gilbert, W. Evolution of the intron-exon structure of eukaryotic genes. Curr. Opin. Genet. Dev. 1995, 5, 774–778. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García, G.C.; Vidriero, I.L.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Tarrío, R.; Ayala, F.J.; Trelles, F.R. Alternative splicing: A missing piece in the puzzle of intron gain. Proc. Natl. Acad. Sci. USA 2008, 105, 7223–7228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.L.; Geisler, M. Genome-Wide Computational Identification of Biologically Significant Cis-Regulatory Elements and Associated Transcription Factors from Rice. Plants 2019, 8, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Zhao, Y.; Jiang, Q.; Yang, J.; Zhao, W.; Taylor, I.A.; Peng, Y.L.; Wang, D.; Liu, J. Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 2019, 47, 4308–4318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarei, A.; Körbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ni, P.Y.; Ji, X.R.; Guo, D.L. Genome-wide identification, characterization, and expression analysis of GDSL-type esterases/lipases gene family in relation to grape berry ripening. Sci. Hortic. 2020, 264, e109162. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Panchy, N.; Shiu, L.M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Omranian, N.; Kleessen, S.; Tohge, T.; Klie, S.; Basler, G.; Roeber, B.M.; Fernie, A.R.; Nikoloski, Z. Differential metabolic and coexpression networks of plant metabolism. Trends Plant Sci. 2015, 20, 266–268. [Google Scholar] [CrossRef]
- Chico, J.M.; Chini, A.; Fonseca, S.; Solano, R. JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 2008, 11, 486–494. [Google Scholar] [CrossRef]
- Gunjegaonkar, S.M.; Shanmugarajan, T.S. Molecular mechanism of plant stress hormone methyl jasmonate for its anti-inflammatory activity. Plant Signal. Behav. 2019, 14, e1642038. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Ding, L.; Yu, H. Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 2013, 32, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. https://doi.org/10.3390/ijms22189974
Sun Y, Liu C, Liu Z, Zhao T, Jiang J, Li J, Xu X, Yang H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. International Journal of Molecular Sciences. 2021; 22(18):9974. https://doi.org/10.3390/ijms22189974
Chicago/Turabian StyleSun, Yaoguang, Chunxin Liu, Zengbing Liu, Tingting Zhao, Jingbin Jiang, Jingfu Li, Xiangyang Xu, and Huanhuan Yang. 2021. "Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato" International Journal of Molecular Sciences 22, no. 18: 9974. https://doi.org/10.3390/ijms22189974




