A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles
Abstract
:1. Introduction
2. Results
2.1. Method Establishment and Validation
2.2. Recovery
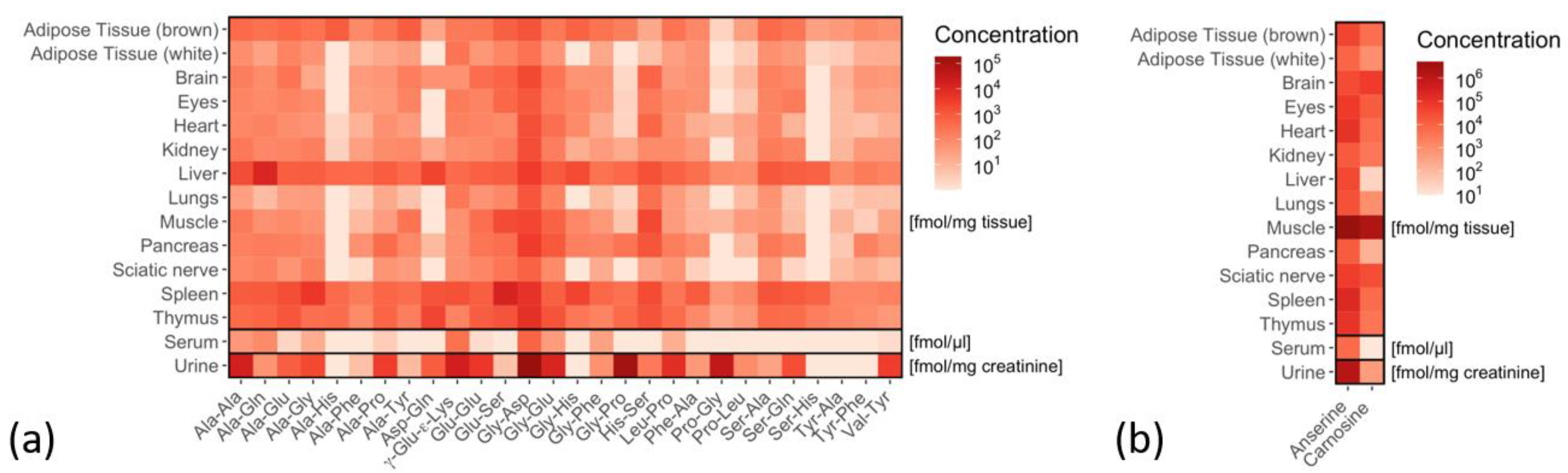
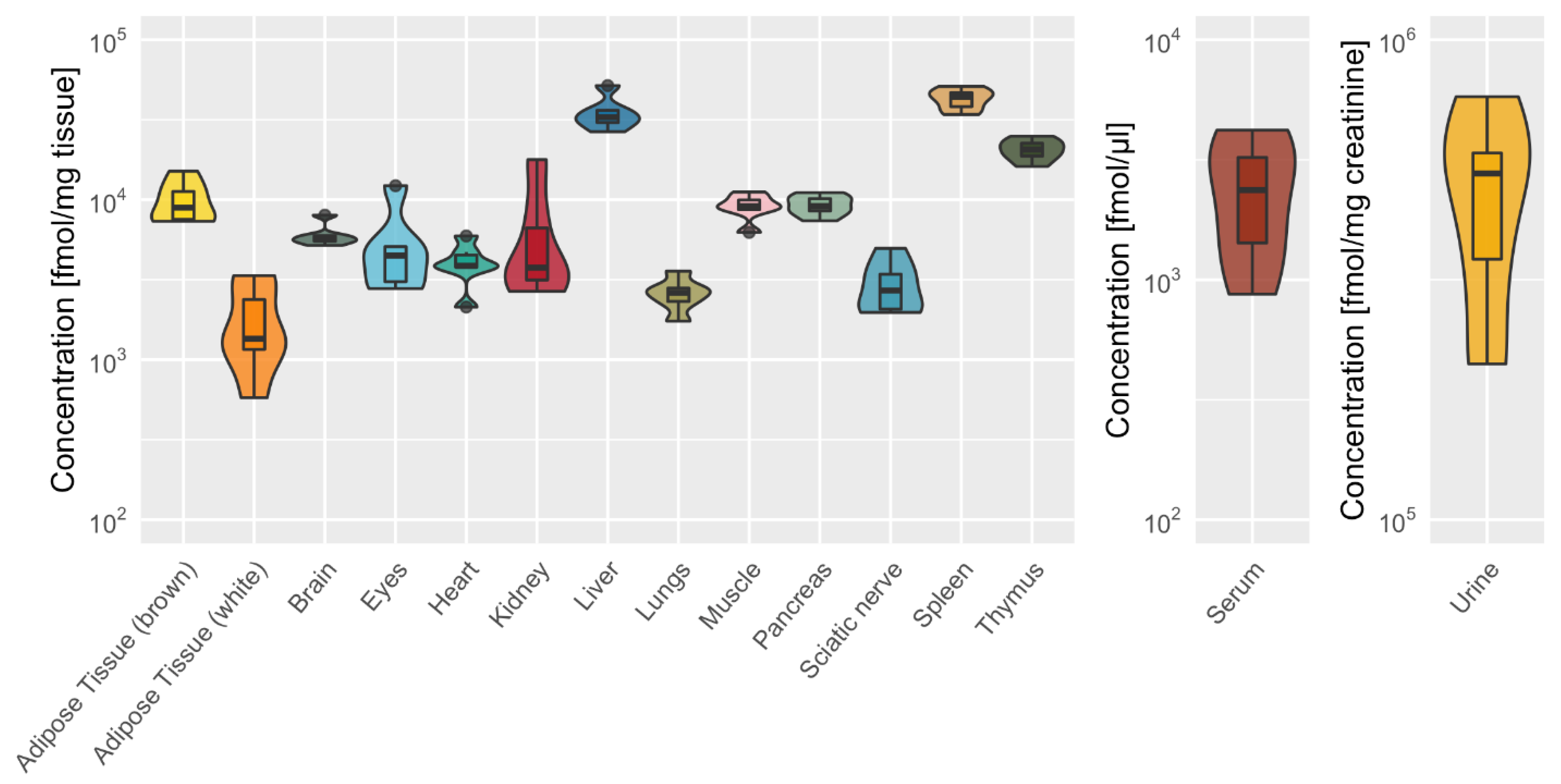
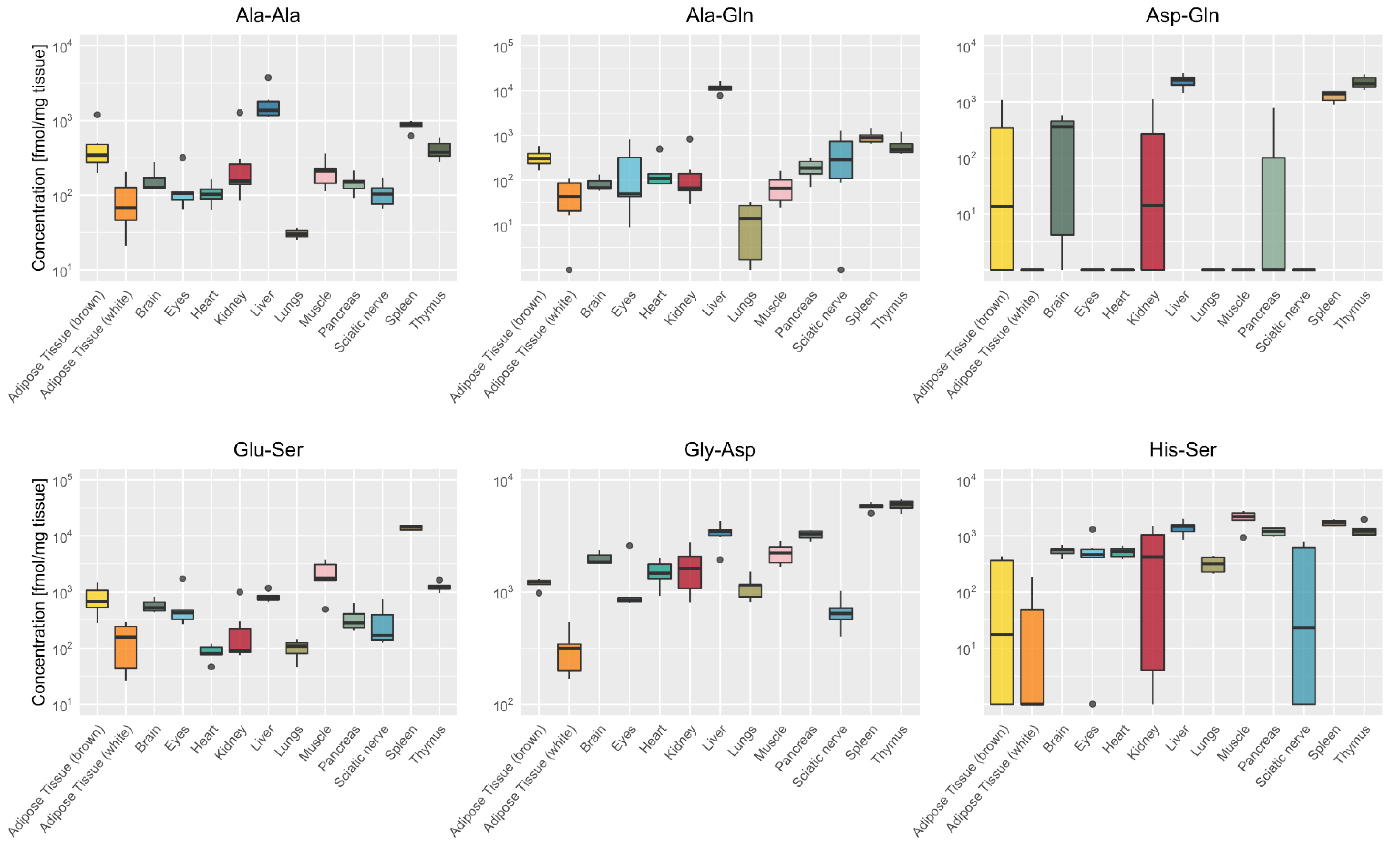
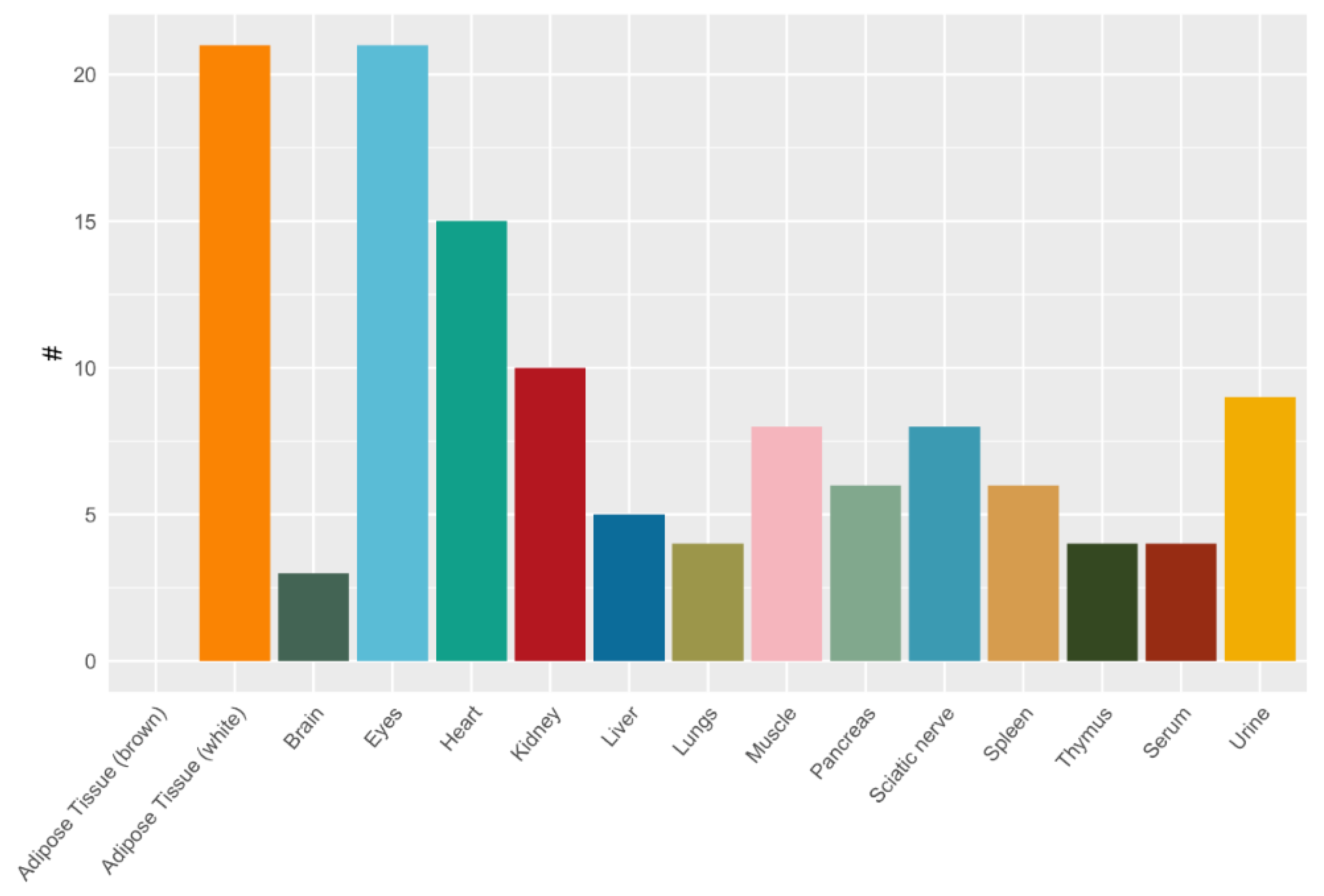
2.3. Tissue-Specific Dipeptide Profiles
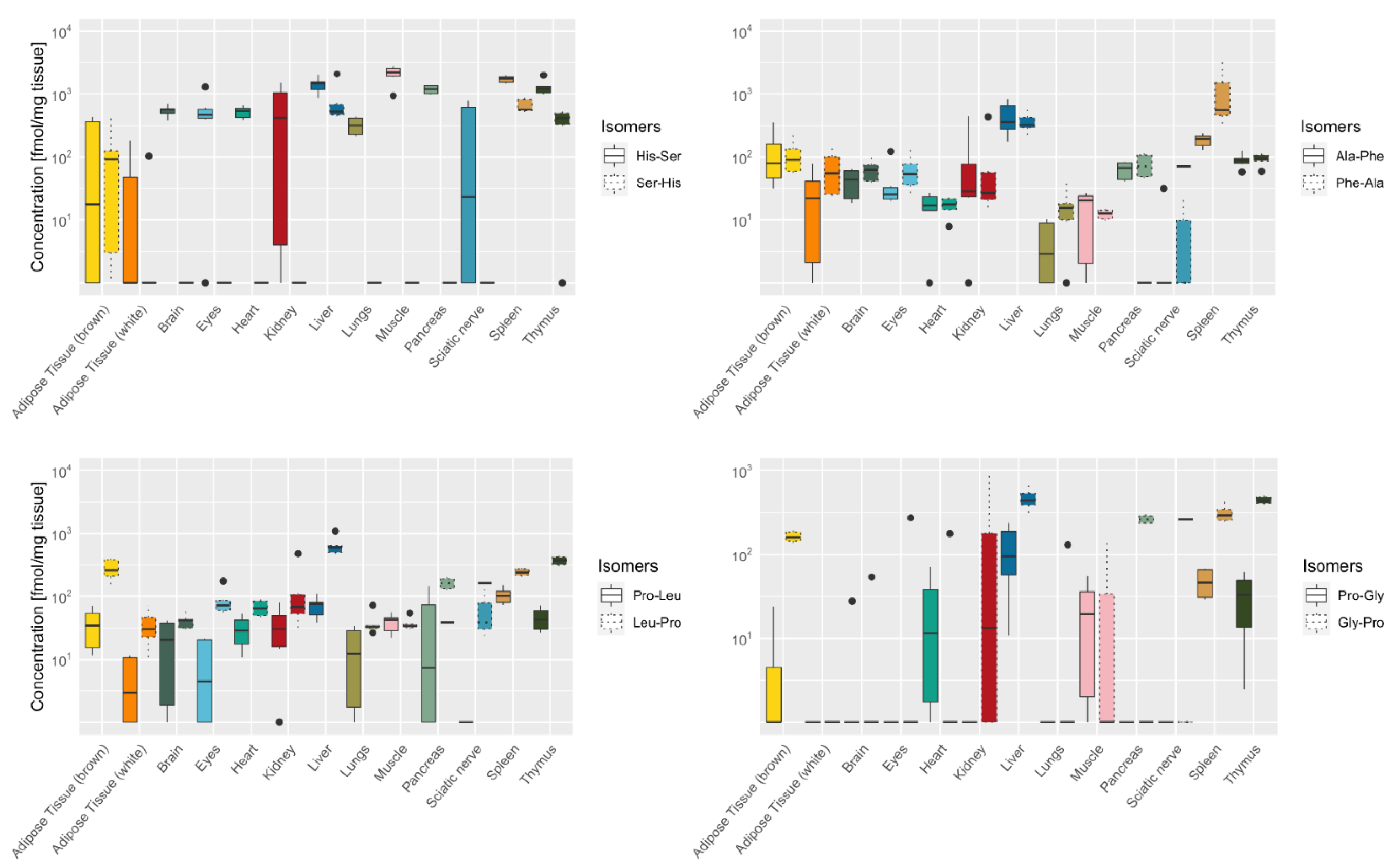
2.4. Tissue-Specific Distribution of Isomeric Dipeptides Forms
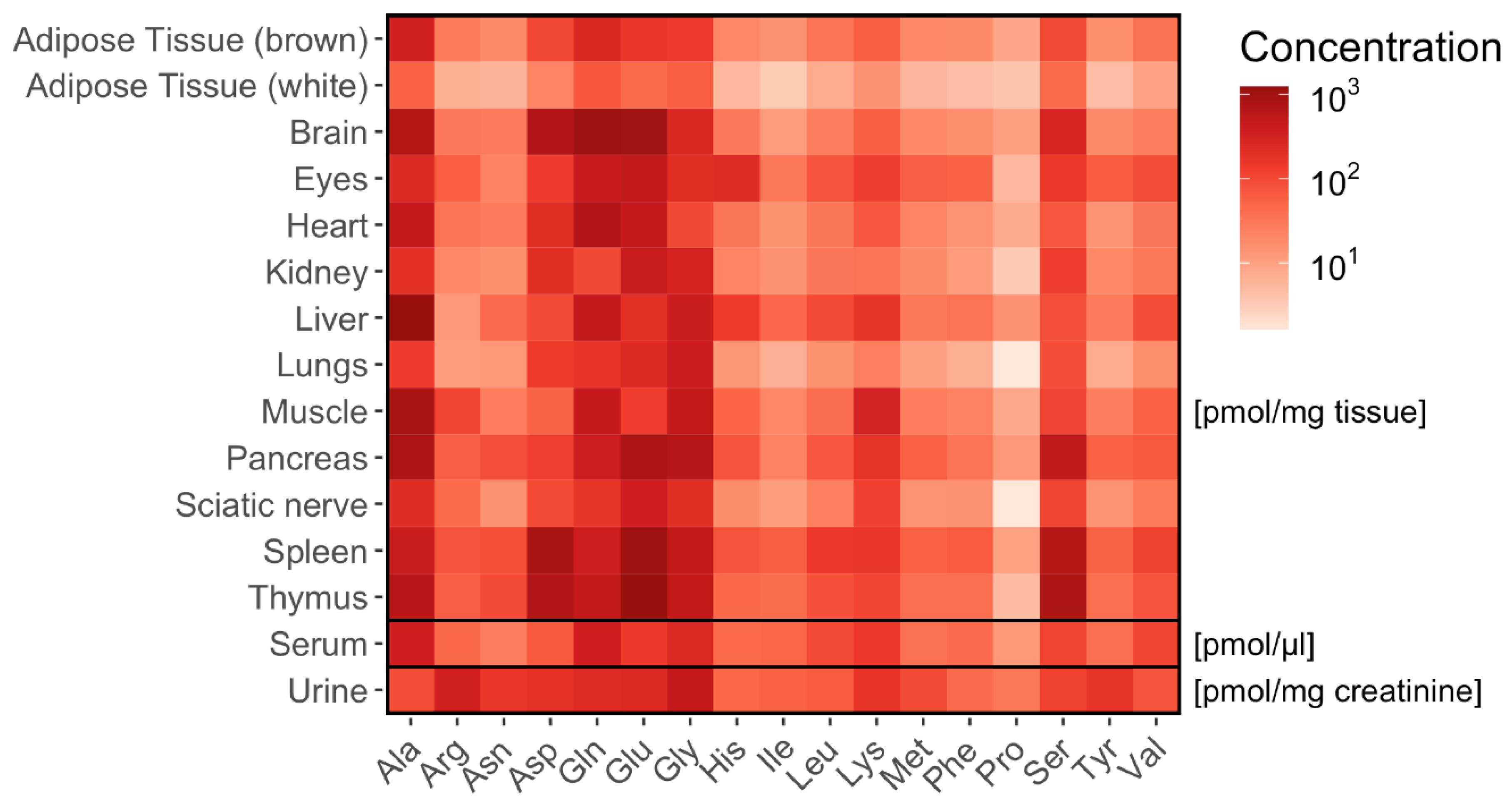
2.5. Tissue-Specific Amino Acid Profiles Relative to Dipeptide Profiles
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Organ Harvest
4.3. Sample Treatment
4.4. Chromatographic Conditions
4.5. Mass Spectrometry Conditions
4.6. Method Validation
4.7. Amino Acid and Creatinine Quantification
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Misiura, M.; Miltyk, W. Proline-containing peptides—New insight and implications: A Review. Biofactors 2019, 45, 857–866. [Google Scholar] [CrossRef]
- Viennois, E.; Pujada, A.; Zen, J.; Merlin, D. Function, regulation, and pathophysiological relevance of the pot superfamily, specifically pept1 in inflammatory bowel disease. Compr. Physiol. 2018, 8, 731–760. [Google Scholar]
- Spanier, B.; Rohm, F. Proton coupled oligopeptide transporter 1 (pept1) function, regulation, and influence on the intestinal homeostasis. Compr. Physiol. 2018, 8, 843–869. [Google Scholar]
- Wang, C.Y.; Liu, S.; Xie, X.N.; Tan, Z.R. Regulation profile of the intestinal peptide transporter 1 (PepT1). Drug Des. Dev. Ther. 2017, 11, 3511–3517. [Google Scholar] [CrossRef] [Green Version]
- Rohm, F.; Daniel, H.; Spanier, B. Transport versus hydrolysis: Reassessing intestinal assimilation of Di- and Tripeptides by LC-MS/MS analysis. Mol. Nutr. Food Res. 2019, 63, e1900263. [Google Scholar] [CrossRef] [Green Version]
- Rohm, F.; Skurk, T.; Daniel, H.; Spanier, B. Appearance of Di- and tripeptides in human plasma after a protein meal does not correlate with pept1 substrate selectivity. Mol. Nutr. Food Res. 2019, 63, e1801094. [Google Scholar] [CrossRef]
- Kwiatkowski, S.; Kiersztan, A.; Drozak, J. Biosynthesis of carnosine and related dipeptides in vertebrates. Curr. Protein Pept. Sci. 2018, 19, 771–789. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef]
- De Koning, T.J. Amino acid synthesis deficiencies. J. Inherit. Metab. Dis. 2017, 40, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armenta, J.M.; Cortes, D.F.; Pisciotta, J.M.; Shuman, J.L.; Blakeslee, K.; Rasoloson, D.; Ogunbiyi, O.; Sullivan, D.J., Jr.; Shulaev, V. Sensitive and rapid method for amino acid quantitation in malaria biological samples using AccQ. Tag ultra performance liquid chromatography-electrospray ionization-MS/MS with multiple reaction monitoring. Anal. Chem. 2010, 82, 548–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casado, M.; Sierra, C.; Batllori, M.; Artuch, R.; Ormazabal, A. A targeted metabolomic procedure for amino acid analysis in different biological specimens by ultra-high-performance liquid chromatography-tandem mass spectrometry. Metabolomics 2018, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Salazar, C.; Armenta, J.M.; Cortés, D.F.; Shulaev, V. Combination of an AccQ·Tag-ultra performance liquid chromatographic method with tandem mass spectrometry for the analysis of amino acids. Methods Mol. Biol. 2012, 828, 13–28. [Google Scholar]
- Weigand, T.; Colbatzky, F.; Pfeffer, T.; Garbade, S.F.; Klingbeil, K.; Colbatzky, F.; Becker, M.; Zemva, J.; Bulkescher, R.; Schürfeld, R.; et al. A global Cndp1-knock-out selectively increases renal carnosine and anserine concentrations in an age- and gender-specific manner in mice. Int. J. Mol. Sci. 2020, 21, 4887. [Google Scholar] [CrossRef]
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef] [PubMed]
- Charlton, M.R. Protein metabolism and liver disease. Baillieres Clin. Endocrinol. Metab. 1996, 10, 617–635. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Hübl, W.; Druml, W.; Roth, E.; Lochs, H. Importance of liver and kidney for the utilization of glutamine-containing dipeptides in man. Metabolism 1994, 43, 1104–1107. [Google Scholar] [CrossRef]
- Langer, G.; Großmann, K.; Fleischer, S.; Berg, A.; Grothues, D.; Wienke, A.; Behrens, J.; Fink, A. Nutritional interventions for liver-transplanted patients. Cochrane Database Syst. Rev. 2012, 8, Cd007605. [Google Scholar] [CrossRef]
- Vermeulen, M.A.; van de Poll, M.C.; Ligthart-Melis, G.C.; Dejong, C.H.; van den Tol, M.P.; Boelens, P.G.; van Leeuwen, P.A. Specific amino acids in the critically ill patient—Exogenous glutamine/arginine: A common denominator? Crit. Care Med. 2007, 35, S568–S576. [Google Scholar] [CrossRef] [PubMed]
- Herzog, R.; Bartosova, M.; Tarantino, S.; Wagner, A.; Unterwurzacher, M.; Sacnun, J.M.; Lichtenauer, A.M.; Kuster, L.; Schaefer, B.; Alper, S.L.; et al. Peritoneal Dialysis fluid supplementation with alanyl-glutamine attenuates conventional dialysis fluid-mediated endothelial cell injury by restoring perturbed cytoprotective responses. Biomolecules 2020, 10, 1678. [Google Scholar] [CrossRef] [PubMed]
- Stehle, P.; Ellger, B.; Kojic, D.; Feuersenger, A.; Schneid, C.; Stover, J.; Scheiner, D.; Westphal, M. Glutamine dipeptide-supplemented parenteral nutrition improves the clinical outcomes of critically ill patients: A systematic evaluation of randomised controlled trials. Clin. Nutr. ESPEN 2017, 17, 75–85. [Google Scholar] [CrossRef]
- Koopman, R.; Caldow, M.K.; Ham, D.J.; Lynch, G.S. Glycine metabolism in skeletal muscle: Implications for metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Gheller, B.J.; Blum, J.E.; Bender, E.L.; Gheller, M.E.; Lim, E.W.; Handzlik, M.K.; Stover, P.J.; Field, M.S.; Cosgrove, B.D.; Metallo, C.M.; et al. Serine and glycine are essential for human muscle progenitor cell population expansion. bioRxiv 2019, 3, 833798. [Google Scholar]
- Martí, I.L.A.A.; Reith, W. Arginine-dependent immune responses. Cell Mol. Life Sci. 2021, 9, 120. [Google Scholar]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus white adipose tissue: A mini-review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Fürst, P. The role of amino acids and synthetic dipeptides. Clin. Nutr. 2003, 22 (Suppl. S2), S23–S28. [Google Scholar] [CrossRef]
- Yagasaki, M.; Hashimoto, S. Synthesis and application of dipeptides; current status and perspectives. Appl. Microbiol. Biotechnol. 2008, 81, 13–22. [Google Scholar] [CrossRef]
- Vistoli, G.; Colzani, M.; Mazzolari, A.; Maddis, D.D.; Grazioso, G.; Pedretti, A.; Carini, M.; Aldini, G. Computational approaches in the rational design of improved carbonyl quenchers: Focus on histidine containing dipeptides. Future Med. Chem. 2016, 8, 1721–1737. [Google Scholar] [CrossRef]
- Peters, V.; Yard, B.; Schmitt, C.P. Carnosine and diabetic nephropathy. Curr. Med. Chem. 2019, 27, 1801–1812. [Google Scholar] [CrossRef]
- Peters, V.; Schmitt, C.P.; Zschocke, J.; Gross, M.L.; Brismar, K.; Forsberg, E. Carnosine treatment largely prevents alterations of renal carnosine metabolism in diabetic mice. Amino Acids 2012, 42, 2411–2416. [Google Scholar] [CrossRef]
- Peters, V.; Calabrese, V.; Forsberg, E.; Volk, N.; Fleming, T.; Baelde, H.; Weigand, T.; Thiel, C.; Trovato, A.; Scuto, M.; et al. Protective actions of anserine under diabetic conditions. Int. J. Mol. Sci. 2018, 19, 2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.C.; Qiao, B.W.; Zhang, Q.; Wang, C.X.; Fu, Y.H.; Zhu, B.W. Study on the domain selective inhibition of angiotensin-converting enzyme (ACE) by food-derived tyrosine-containing dipeptides. J. Food Biochem. 2021, 45, e13779. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.C.; Rojas, B.E.; Vicente, R.; Gorka, M.; Matz, T.; Chodasiewicz, M.; Peralta-Ariza, J.S.; Zhang, Y.; Alseekh, S.; Childs, D.; et al. Tyr-Asp inhibition of glyceraldehyde 3-phosphate dehydrogenase affects plant redox metabolism. EMBO J. 2021, 40, e106800. [Google Scholar] [CrossRef]
- Moravek, M.; Fisseha, S.; Swain, J.E. Dipeptide forms of glycine support mouse preimplantation embryo development in vitro and provide protection against high media osmolality. J. Assist. Reprod. Genet. 2012, 29, 283–290. [Google Scholar] [CrossRef] [Green Version]
- El Nahas, A.M.; Abo-Zenah, H.; Skill, N.J.; Bex, S.; Wild, G.; Griffin, M.; Johnson, T.S. Elevated epsilon-(gamma-glutamyl) lysine in human diabetic nephropathy results from increased expression and cellular release of tissue transglutaminase. Nephron Clin. Pract. 2004, 97, c108–c117. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, S.N.S.; Sciani, J.M.; Lisboa, N.M.; Stefani, H.A. Synthesis of a tyr-tyr dipeptide library and evaluation against tumor cells. Med. Chem. 2018, 14, 709–714. [Google Scholar] [CrossRef]
- Ano, Y.; Yoshino, Y.; Uchida, K.; Nakayama, H. Preventive effects of tryptophan-methionine dipeptide on neural inflammation and Alzheimer’s pathology. Int. J. Mol. Sci. 2019, 20, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeitner, T.M.; Matson, W.R.; Folk, J.E.; Blass, J.P.; Cooper, A.J. Increased levels of gamma-glutamylamines in Huntington disease CSF. J. Neurochem. 2008, 106, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mižíková, I.; Pfeffer, T.; Nardiello, C.; Surate Solaligue, D.E.; Steenbock, H.; Tatsukawa, H.; Silva, D.M.; Vadász, I.; Herold, S.; Pease, R.J.; et al. Targeting transglutaminase 2 partially restores extracellular matrix structure but not alveolar architecture in experimental bronchopulmonary dysplasia. FEBS J. 2018, 285, 3056–3076. [Google Scholar] [CrossRef]
- Gu, X.; Al Dubayee, M.; Alshahrani, A.; Masood, A.; Benabdelkamel, H.; Zahra, M.; Li, L.; Abdel Rahman, A.M.; Aljada, A. Distinctive metabolomics patterns associated with insulin resistance and type 2 diabetes mellitus. Front. Mol. Biosci. 2020, 7, 609806. [Google Scholar] [CrossRef]
- Wu, G. Dietary protein intake and human health. Food Funct. 2016, 7, 1251–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weger, B.D.; Weger, M.; Görling, B.; Schink, A.; Gobet, C.; Keime, C.; Poschet, G.; Jost, B.; Krone, N.; Hell, R.; et al. Extensive regulation of diurnal transcription and metabolism by glucocorticoids. PLoS Genet. 2016, 12, e1006512. [Google Scholar] [CrossRef] [PubMed]
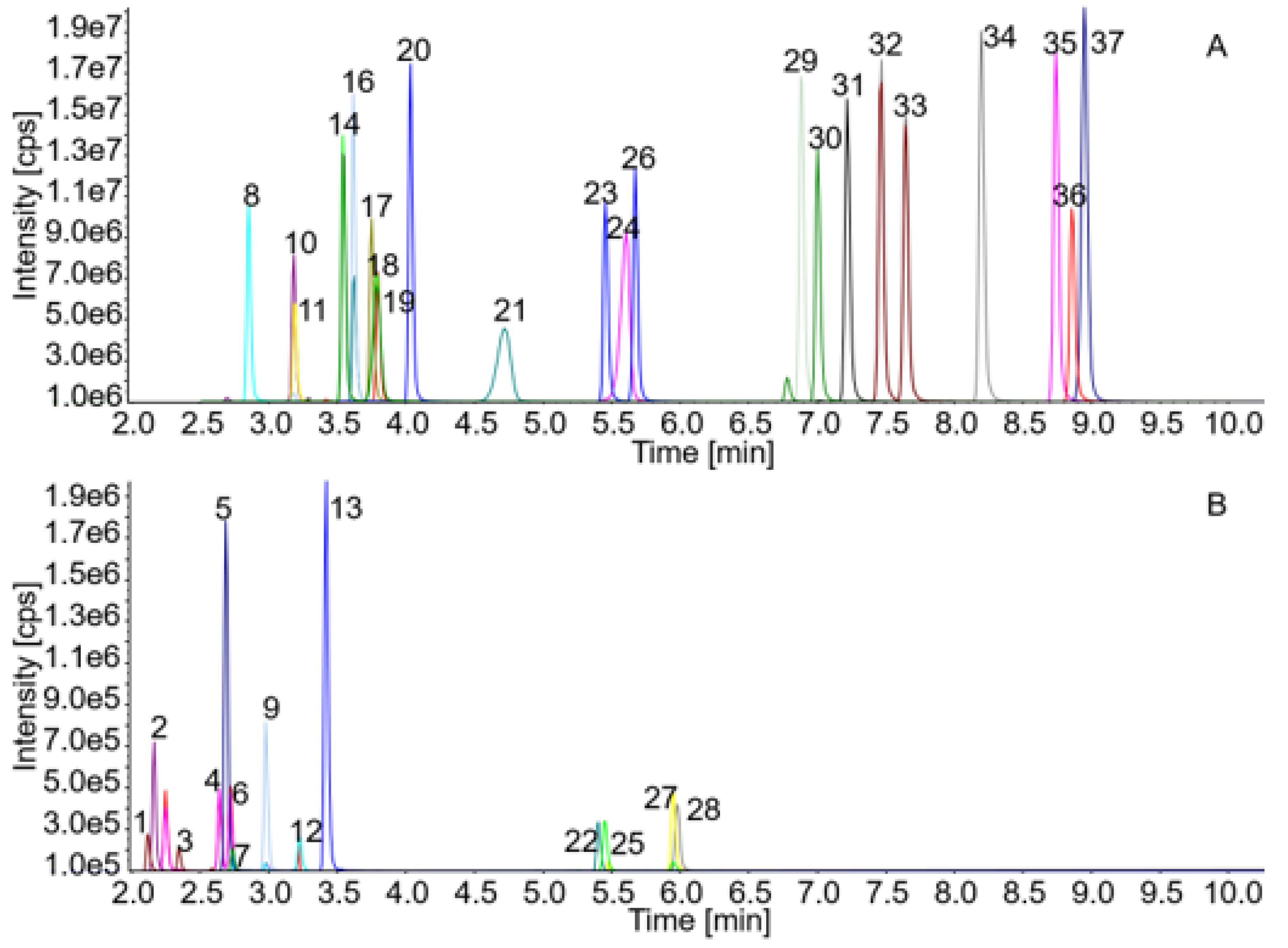

| Peak No. | Analyte | Q1 | Q3 | RT | DP | CE | CXP |
|---|---|---|---|---|---|---|---|
| 1 | Seryl-Histidine (Ser-His) | 413.0 | 171.1 | 2.08 | 61 | 39 | 20 |
| 2 | Glycyl-Histidine (Gly-His) | 383.0 | 213.1 | 2.15 | 31 | 21 | 12 |
| 171.1 | 35 | 20 | |||||
| 3 | Histidyl-Serine (His-Ser) | 413.0 | 171.1 | 2.31 | 46 | 31 | 16 |
| 4 | Carnosine (β-Ala-His) | 397.0 | 171.1 | 2.62 | 56 | 43 | 18 |
| 110 | 51 | 14 | |||||
| 5 | Serinyl-Glutamine (Ser-Gln) | 403.9 | 171.1 | 2.65 | 126 | 53 | 10 |
| 6 | Alanyl-Histidine (Ala-His) | 397.0 | 171.1 | 2.71 | 46 | 35 | 20 |
| 7 | Anserine (β-Ala-3methyl-His) | 411.0 | 171.1 | 2.73 | 131 | 39 | 10 |
| 8 | Glycyl-Aspartic Acid (Gly-Asp) | 361.0 | 171.1 | 2.84 | 31 | 31 | 16 |
| 9 | Aspartyl-Glutamine (Asp-Gln) | 431.9 | 171.1 | 2.97 | 46 | 27 | 18 |
| 10 | Glutamyl-Serine (Glu-Ser) | 404.9 | 171.1 | 3.16 | 131 | 35 | 20 |
| 11 | Glycyl-Glutamic Acid (Gly-Glu) | 374.9 | 171.1 | 3.18 | 146 | 25 | 10 |
| 12 | Histidylalanine (His-Ala) | 397.0 | 171.1 | 3.23 | 51 | 31 | 14 |
| 13 | Alanyl-Glutamine (Ala-Gln) | 387.9 | 171.1 | 3.41 | 106 | 27 | 14 |
| 14 | Alanyl-Glycine (Ala-Gly) | 317.0 | 171.1 | 3.53 | 31 | 33 | 12 |
| 15 | Seryl-Alanine (Ser-Ala) | 347.0 | 171.1 | 3.6 | 51 | 33 | 18 |
| 16 | Glutamyl-Glutamic Acid (Glu-Glu) | 447.0 | 171.1 | 3.61 | 41 | 41 | 18 |
| 17 | Prolyl-Glycine (Pro-Gly) | 343.0 | 171.1 | 3.73 | 66 | 35 | 20 |
| 89 | 103 | 10 | |||||
| 18 | Glycyl-Sarcosine (Gly-Sar) | 317.0 | 171.1 | 3.77 | 56 | 31 | 20 |
| 116 | 77 | 12 | |||||
| 19 | Alanyl-Glutamic Acid (Ala-Glu) | 388.9 | 171.1 | 3.79 | 126 | 23 | 10 |
| 20 | Alanyl-Alanine (Ala-Ala) | 331.0 | 171.1 | 4.05 | 36 | 31 | 10 |
| 21 | Glycyl-Proline (Gly-Pro) | 343.0 | 171.1 | 4.71 | 86 | 31 | 20 |
| 22 | γ-Glutamyl-ε-Lysine (γ-Glu-ε-Lys) | 616.0 | 171.1 | 5.41 | 31 | 49 | 12 |
| 23 | Alanyl-Tyrosine (Ala-Tyr) | 422.9 | 171.1 | 5.46 | 21 | 37 | 20 |
| 24 | Alanyl-Proline (Ala-Pro) | 357.0 | 171.1 | 5.61 | 36 | 33 | 20 |
| 25 | Leucyl-Histidine (Leu-His) | 439.0 | 156 | 5.64 | 26 | 33 | 18 |
| 171.1 | 33 | 18 | |||||
| 26 | Tyrosyl-Alanine (Tyr-Ala) | 422.9 | 171.1 | 5.65 | 21 | 39 | 20 |
| 27 | Histidyl-Leucine (His-Leu) | 439.1 | 110 | 5.97 | 26 | 43 | 12 |
| 171.1 | 31 | 10 | |||||
| 28 | Arginyl-Phenylalanine (Arg-Phe) | 492.1 | 171.1 | 6 | 36 | 47 | 20 |
| 29 | Glycyl-Phenylalanine (Gly-Phe) | 392.9 | 171.1 | 6.98 | 16 | 37 | 20 |
| 30 | Valyl-Tyrosine (Val-Tyr) | 451.0 | 171.1 | 7 | 26 | 39 | 16 |
| 31 | Prolyl-Leucine (Pro-Leu) | 399.1 | 171 | 7.22 | 46 | 39 | 20 |
| 229.2 | 23 | 14 | |||||
| 32 | Alanyl-Phenylalanine (Ala-Phe) | 407.0 | 171.1 | 7.48 | 56 | 37 | 20 |
| 242.1 | 21 | 16 | |||||
| 33 | Phenylalanyl-Alanine (Phe-Ala) | 407.0 | 171.1 | 7.66 | 61 | 37 | 10 |
| 120.1 | 29 | 14 | |||||
| 34 | Internal Std—Norleucine (2-Aminohexanoic acid) | 302.0 | 171.1 | 8.21 | 81 | 25 | 16 |
| 35 | Aspartame (Asp-Phe-methylester) | 465.1 | 171.1 | 8.77 | 21 | 37 | 20 |
| 36 | Tyrosyl-Phenylalanine (Tyr-Phe) | 499.0 | 171.1 | 8.86 | 26 | 41 | 20 |
| 37 | Leucyl-Proline (Leu-Pro) | 399.1 | 171.1 | 8.97 | 56 | 37 | 20 |
| 229.2 | 21 | 20 |
| Time (min) | Flow (mL/min) | %A | %B |
|---|---|---|---|
| Initial | 0.5 | 98 | 2 |
| 0.50 | 0.5 | 98 | 2 |
| 11.50 | 0.5 | 70 | 30 |
| 11.51 | 0.5 | 10 | 90 |
| 13.00 | 0.5 | 10 | 90 |
| 13.10 | 0.5 | 98 | 2 |
| 16.00 | 0.5 | 98 | 2 |
| Parameter | Details |
|---|---|
| Scan type | MRM |
| Polarity | positive |
| MRM detection window (s) | 60 |
| Target scan time per sMRM (s) | 0.6 |
| Curtain Gas (CUR) | 40 |
| Collision Gas | Medium |
| IonSpray Voltage (IS) | 5500 |
| Temperature (TEM) | 550 |
| Ion Source Gas 1 (GS1) | 70 |
| Ion Source Gas 2 (GS2) | 70 |
| Entrance Potential (EP) | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heidenreich, E.; Pfeffer, T.; Kracke, T.; Mechtel, N.; Nawroth, P.; Hoffmann, G.F.; Schmitt, C.P.; Hell, R.; Poschet, G.; Peters, V. A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles. Int. J. Mol. Sci. 2021, 22, 9979. https://doi.org/10.3390/ijms22189979
Heidenreich E, Pfeffer T, Kracke T, Mechtel N, Nawroth P, Hoffmann GF, Schmitt CP, Hell R, Poschet G, Peters V. A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles. International Journal of Molecular Sciences. 2021; 22(18):9979. https://doi.org/10.3390/ijms22189979
Chicago/Turabian StyleHeidenreich, Elena, Tilman Pfeffer, Tamara Kracke, Nils Mechtel, Peter Nawroth, Georg F Hoffmann, Claus Peter Schmitt, Rüdiger Hell, Gernot Poschet, and Verena Peters. 2021. "A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles" International Journal of Molecular Sciences 22, no. 18: 9979. https://doi.org/10.3390/ijms22189979
APA StyleHeidenreich, E., Pfeffer, T., Kracke, T., Mechtel, N., Nawroth, P., Hoffmann, G. F., Schmitt, C. P., Hell, R., Poschet, G., & Peters, V. (2021). A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles. International Journal of Molecular Sciences, 22(18), 9979. https://doi.org/10.3390/ijms22189979






