Emerging Functions for snoRNAs and snoRNA-Derived Fragments
Abstract
:1. Introduction
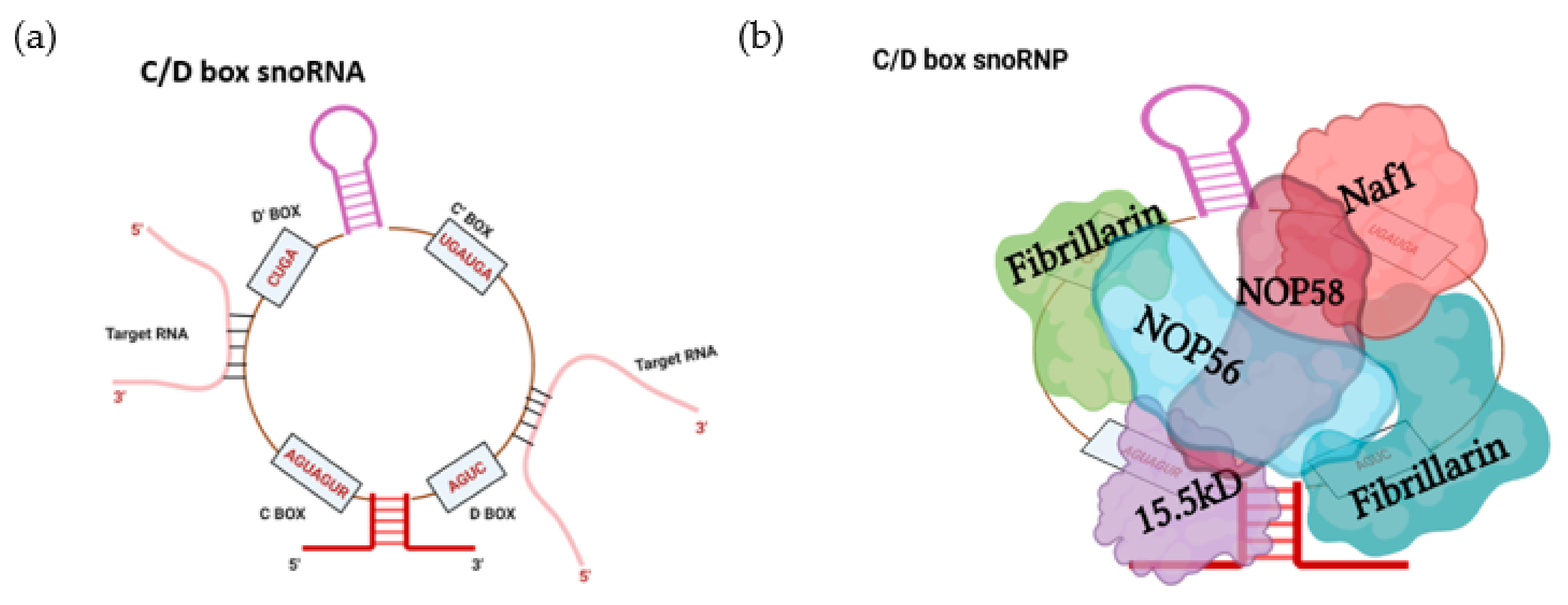
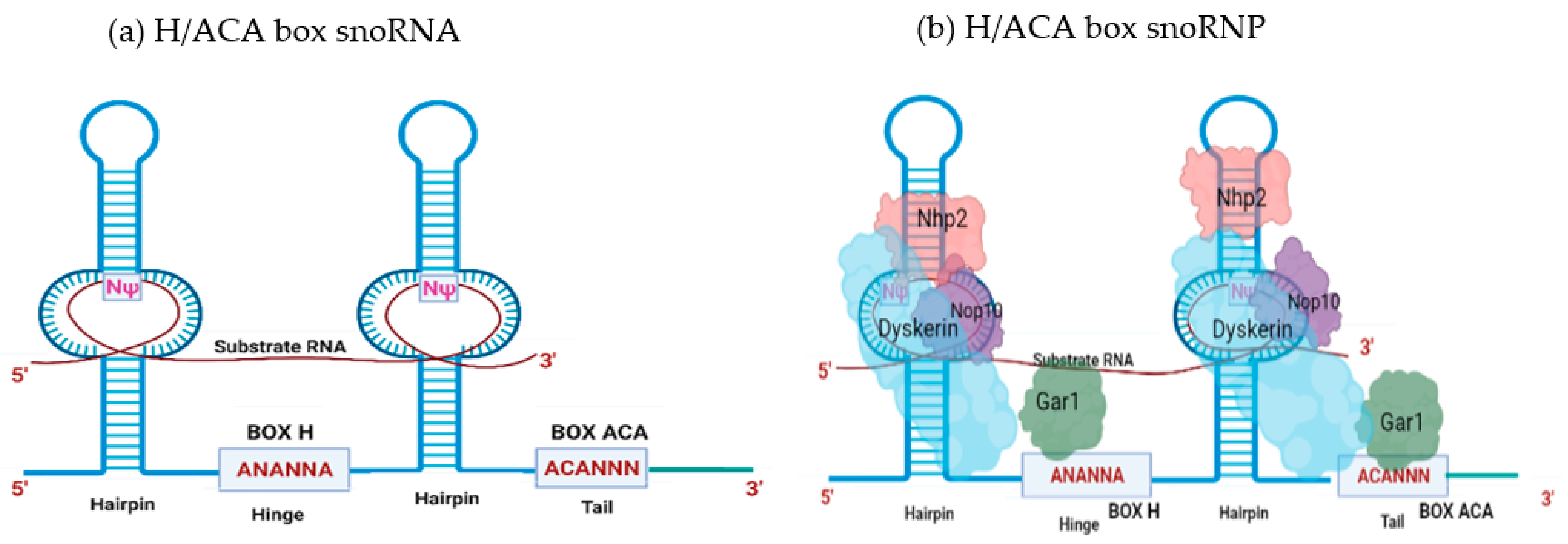
2. SnoRNA Classes and Canonical Function
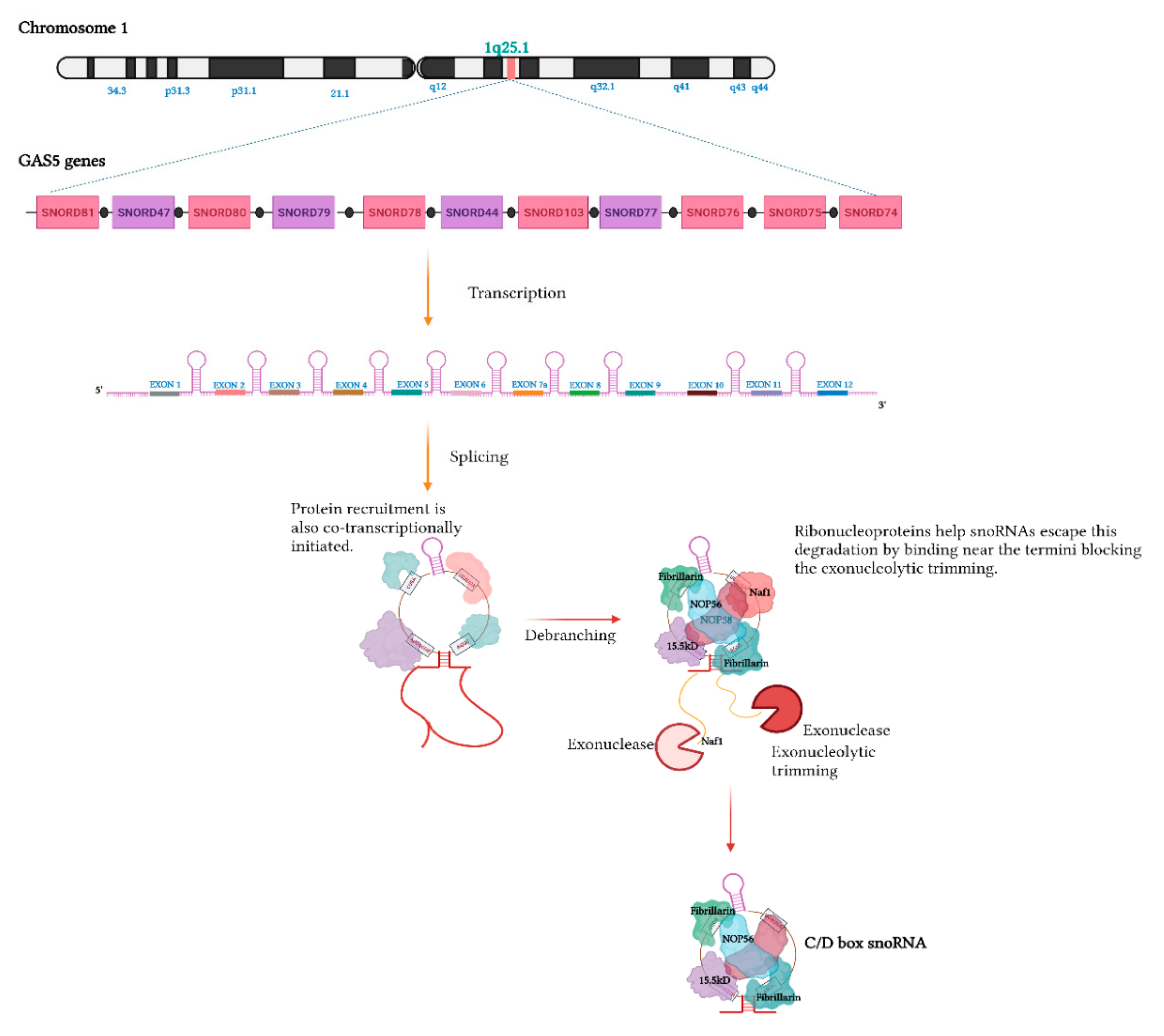
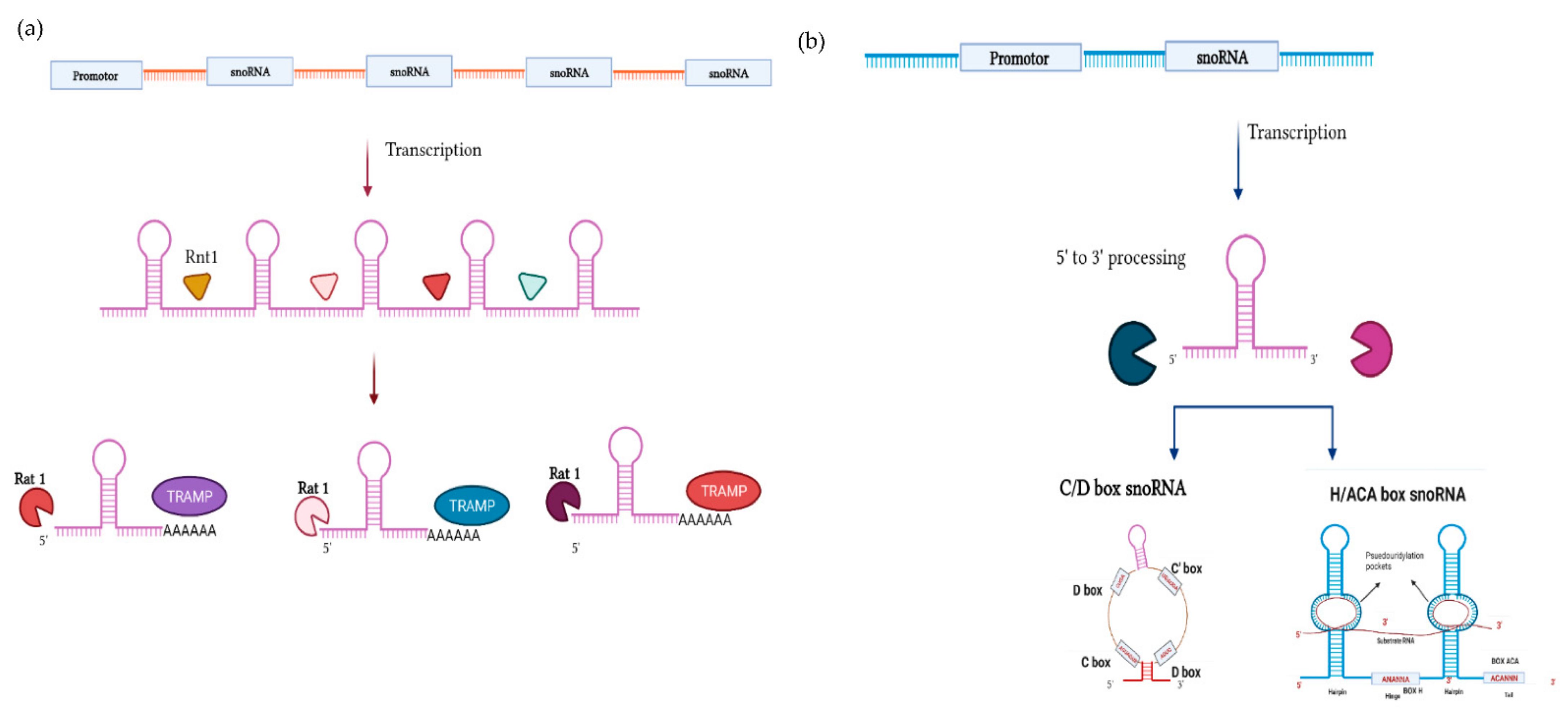
Biogenesis
3. Diversification of ncRNA Function
3.1. miRNAs
3.2. tRNAs and tRFs
3.3. SnoRNAs—Mechanistic Roles in Cancer and Their Capacity as Biomarkers
4. Emerging snoRNA Non-Canonical Functions
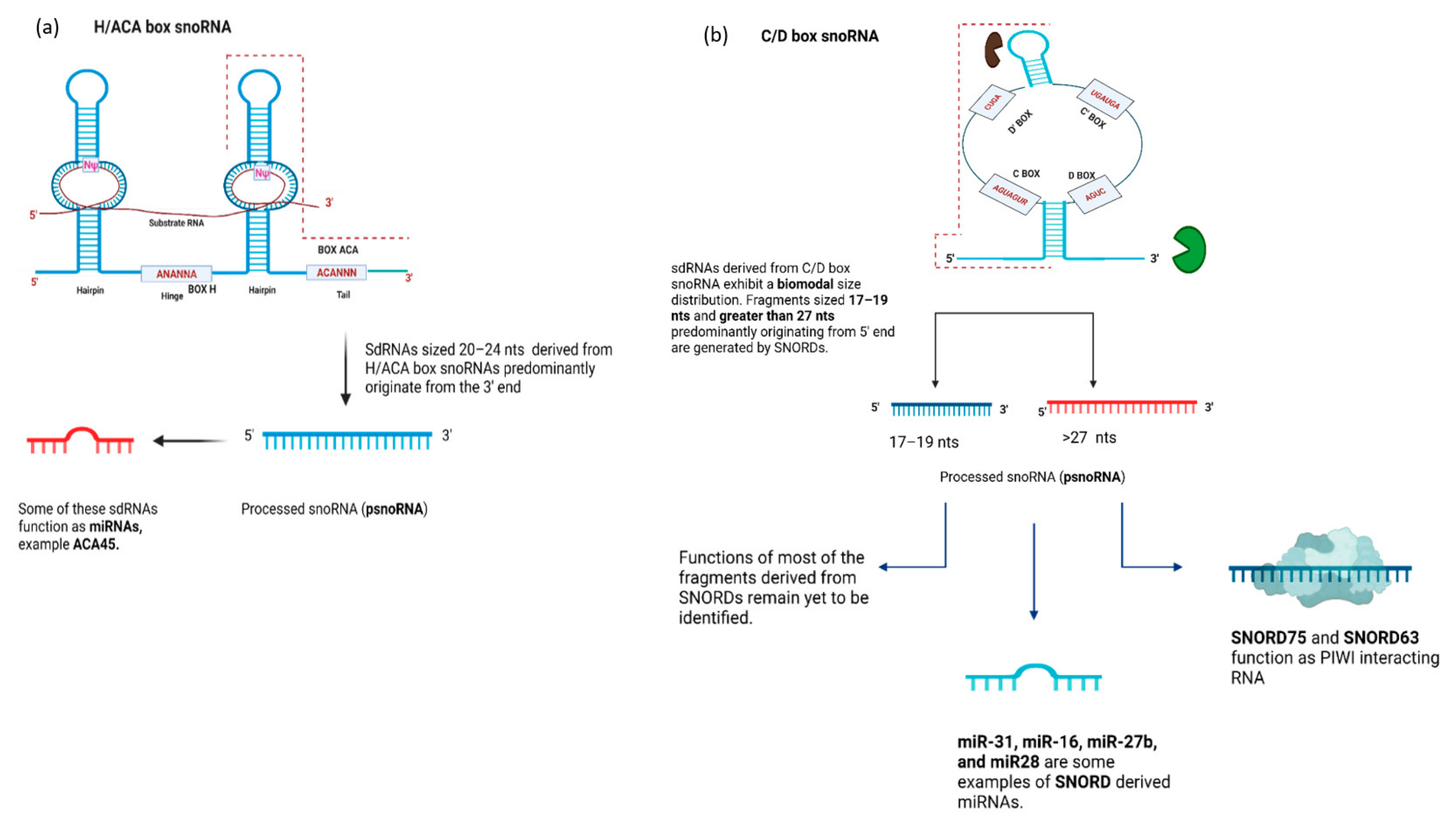
4.1. snoRNA-Derived RNAs (sdRNAs)—Function
4.2. SnoRNA-Derived RNAs (sdRNAs)—Biogenesis
5. Conclusions
6. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviations | Meaning |
| ncRNAs | Non-coding RNAs |
| rRNA | Ribosomal RNA |
| tRNA | Transfer RNA |
| miRNA | microRNA |
| piRNA | PIWI-interacting RNA |
| siRNA | Small interfering RNA |
| lncRNA | Long non-coding RNA |
| paRNAs | Promoter-associated RNAs |
| SnoRNA | Small nucleolar RNA |
| sdRNA | snoRNA-derived fragments |
| snRNA | Small nuclear RNAs |
| snoRNP | snoRNA–ribonucleoprotein complexes |
| scaRNAs | Small Cajal body specific RNAs |
| RISC | RNA-induced silencing complex |
| RITS | RNA-induced transcriptional silencing1 |
| RTD | Rapid tRNA decay |
| tRFs | tRNA-derived fragments |
| ANG | Angiogenin |
| AGO | Argonaute |
| tiRNAs | Stress-induced tRNA fragments |
| tRF-GG | tRF-Gly-GCC |
| ERV | Endogenous retroviral element |
| HCC | Hepatocellular carcinoma |
| CRCs | Colorectal carcinomas |
| ALDH | Aldehyde dehydrogenase |
| NSCLC | Non-small cell lung cancer |
| Htr2c | Serotonin receptor 2c |
| A–I | Adenosine-to-inosine |
| FGFR3 | Fibroblast growth factor receptor |
| CPSF | Cleavage and polyadenylation specificity factor |
| HEK293 | Human embryonic kidney 293 cells |
| ACA45 | H/ACA scaRNA15 |
| IL-4 | Interleukin-4 |
| TRAMP | Trf4-Air2-Mtr4 |
| FUS | Fused in sarcoma/translocated in liposarcoma |
| hnRNPs | Heterogeneous nuclear ribonucleoprotein |
References
- Hoagland, M.B.; Stephenson, M.L.; Scott, J.F.; Hecht, L.I.; Zamecnik, P.C. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958, 231, 241–257. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Liang, X.-h.; Liu, Q.; Liu, Q.; King, T.H.; Fournier, M.J. Strong dependence between functional domains in a dual-function snoRNA infers coupling of rRNA processing and modification events. Nucleic Acids Res. 2010, 38, 3376–3387. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, L.B.; Steitz, J.A. Guided tours: From precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999, 11, 378–384. [Google Scholar] [CrossRef]
- Maxwell, E.; Fournier, M. The small nucleolar RNAs. Annu. Rev. Biochem. 1995, 64, 897–934. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Consortium, M.G.S.; Waterston, R.; Lindblad-Toh, K.; Birney, E.; Rogers, J. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar]
- Consortium, I. A haplotype map of the human genome. Nature 2005, 437, 1299–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [Green Version]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [Green Version]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.X.; Ma, J.X. Promoter-associated RNAs and promoter-targeted RNAs. Cell. Mol. Life Sci. 2012, 69, 2833–2842. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Scott, M.S.; Yamada, K.; Avolio, F.; Barton, G.J.; Lamond, A.I. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011, 39, 3879–3891. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, X.; Zhang, X.; Duan, X.; Pan, T.; Hu, Q.; Zhang, Y.; Zhong, F.; Liu, J.; Zhang, H. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015, 43, 3712–3725. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Olvedy, M.; Jenster, G. Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013, 340, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Tirasophon, W.; Roshorm, Y.; Panyim, S. Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem. Biophys. Res. Commun. 2005, 334, 102–107. [Google Scholar] [CrossRef]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhang, X.; Shi, J.; Yan, M.; Zhou, T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.P.; Hutvagner, G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life 2015, 5, 1638–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Krell, J.; Frampton, A.E.; Mirnezami, R.; Harding, V.; De Giorgio, A.; Alonso, L.R.; Cohen, P.; Ottaviani, S.; Colombo, T.; Jacob, J. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS ONE 2014, 9, e98561. [Google Scholar] [CrossRef] [Green Version]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, E.; Fournier, M.J. The snoRNPs and related machines: Ancient devices that mediate maturation of rRNA and other RNAs. In Madame Curie Bioscience Database [Internet]; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Busch, H.; Reddy, R.; Rothblum, L.; Choi, Y. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982, 51, 617–654. [Google Scholar] [CrossRef] [PubMed]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef] [Green Version]
- Reichow, S.L.; Hamma, T.; Ferré-D’Amaré, A.R.; Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001, 20, 3617–3622. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [Green Version]
- Bachellerie, J.-P.; Cavaillé, J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997, 22, 257–261. [Google Scholar] [CrossRef]
- Omer, A.D.; Lowe, T.M.; Russell, A.G.; Ebhardt, H.; Eddy, S.R.; Dennis, P.P. Homologs of Small Nucleolar RNAs in Archaea. Science 2000, 288, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Kiss-László, Z.; Henry, Y.; Kiss, T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998, 17, 797–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, N.J.; Ségault, V.; Charpentier, B.; Nottrott, S.; Fabrizio, P.; Bachi, A.; Wilm, M.; Rosbash, M.; Branlant, C.; Lührmann, R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 2000, 103, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2019, 48, 1627–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peculis, B.A. RNA-binding proteins: If it looks like a sn(o)RNA. Curr. Biol. 2000, 10, R916–R918. [Google Scholar] [CrossRef] [Green Version]
- Méreau, A.; Fournier, R.; Grégoire, A.; Mougin, A.; Fabrizio, P.; Lührmann, R.; Branlant, C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: Protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997, 273, 552–571. [Google Scholar] [CrossRef]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 2013, 35, 46–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakin, A.G.; Smith, L.; Fournier, M.J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 1996, 86, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Dragon, F.; Lemay, V.; Trahan, C. snoRNAs: Biogenesis, structure and function. e LS 2001. [Google Scholar]
- Terns, M.; Terns, R. Noncoding RNAs of the H/ACA family. In Proceedings of the Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2006; pp. 395–405. [Google Scholar]
- Kiss, T.; Fayet-Lebaron, E.; Jády, B.E. Box H/ACA small ribonucleoproteins. Mol. Cell 2010, 37, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Meier, U.T. RNA modification in Cajal bodies. RNA Biol. 2017, 14, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Ojha, S.; Malla, S.; Lyons, S.M. snoRNPs: Functions in Ribosome Biogenesis. Biomolecules 2020, 10, 783. [Google Scholar] [CrossRef]
- Fournier, M.J.; Stuart Maxwell, E. The nucleolar snRNAs: Catching up with the spliceosomal snRNAs. Trends Biochem. Sci. 1993, 18, 131–135. [Google Scholar] [CrossRef]
- Enright, C.A.; Maxwell, E.S.; Eliceiri, G.; Sollner-Webb, B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 1996, 2, 1094–1099. [Google Scholar]
- Atzorn, V.; Fragapane, P.; Kiss, T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004, 24, 1769–1778. [Google Scholar] [CrossRef] [Green Version]
- Tycowski, K.T.; Shu, M.-D.; Steitz, J.A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 1994, 266, 1558–1561. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.W.; Marshall, D.F.; Echeverria, M. Intronic noncoding RNAs and splicing. Trends Plant. Sci. 2008, 13, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Grzechnik, P. Small nucleolar RNAs tell a different tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimta, A.A.; Tigu, A.B.; Braicu, C.; Stefan, C.; Ionescu, C.; Berindan-Neagoe, I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 2020, 10, 389. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Dupuis-Sandoval, F.; Poirier, M.; Scott, M.S. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 2015, 6, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Warf, M.B.; Berglund, J.A. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA 2007, 13, 2238–2251. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.M.; Steitz, J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998, 18, 6897–6909. [Google Scholar] [CrossRef] [Green Version]
- Leader, D.J.; Clark, G.P.; Watters, J.; Beven, A.F.; Shaw, P.J.; Brown, J.W. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997, 16, 5742–5751. [Google Scholar] [CrossRef] [Green Version]
- van Hoof, A.; Lennertz, P.; Parker, R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000, 20, 441–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzechnik, P.; Kufel, J. Polyadenylation Linked to Transcription Termination Directs the Processing of snoRNA Precursors in Yeast. Mol. Cell 2008, 32, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Grzechnik, P.; Szczepaniak, S.A.; Dhir, S.; Pastucha, A.; Parslow, H.; Matuszek, Z.; Mischo, H.E.; Kufel, J.; Proudfoot, N.J. Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage. Nat. Commun. 2018, 9, 1783. [Google Scholar] [CrossRef] [Green Version]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Dalla Venezia, N.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.-J. Emerging role of eukaryote ribosomes in translational control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef] [Green Version]
- Guglas, K.; Kołodziejczak, I.; Kolenda, T.; Kopczyńska, M.; Teresiak, A.; Sobocińska, J.; Bliźniak, R.; Lamperska, K. YRNAs and YRNA-Derived Fragments as New Players in Cancer Research and Their Potential Role in Diagnostics. Int. J. Mol. Sci. 2020, 21, 5682. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Bellodi, C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020, 17, 1214–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Metazoan micrornas. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef] [Green Version]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, M.S.; Avolio, F.; Ono, M.; Lamond, A.I.; Barton, G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009, 5, e1000507. [Google Scholar] [CrossRef] [Green Version]
- Saraiya, A.A.; Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef] [Green Version]
- Turunen, T.A.; Roberts, T.C.; Laitinen, P.; Väänänen, M.-A.; Korhonen, P.; Malm, T.; Ylä-Herttuala, S.; Turunen, M.P. Changes in nuclear and cytoplasmic microRNA distribution in response to hypoxic stress. Sci. Rep. 2019, 9, 10332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, H.W.; Wentzel, E.A.; Mendell, J.T. A hexanucleotide element directs microRNA nuclear import. Science 2007, 315, 97–100. [Google Scholar] [CrossRef]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE 2010, 5, e10563. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e188. [Google Scholar] [CrossRef]
- Younger, S.T.; Pertsemlidis, A.; Corey, D.R. Predicting potential miRNA target sites within gene promoters. Bioorg. Med. Chem. Lett. 2009, 19, 3791–3794. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Sætrom, P.; Snøve, O.; Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, M.; Chu, Y.; Zhang, H.; Gagnon, K.T.; Shaikh, S.; Kuchimanchi, S.; Manoharan, M.; Corey, D.R.; Janowski, B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013, 41, 10086–10109. [Google Scholar] [CrossRef]
- Han, J.; Kim, D.; Morris, K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. Sci. USA 2007, 104, 12422–12427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, J.C.; Younger, S.T.; Nguyen, N.-B.; Hardy, D.B.; Monia, B.P.; Corey, D.R.; Janowski, B.A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008, 15, 842. [Google Scholar] [CrossRef] [PubMed]
- Alló, M.; Buggiano, V.; Fededa, J.P.; Petrillo, E.; Schor, I.; De La Mata, M.; Agirre, E.; Plass, M.; Eyras, E.; Abou Elela, S. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 717. [Google Scholar] [CrossRef] [PubMed]
- Ameyar-Zazoua, M.; Rachez, C.; Souidi, M.; Robin, P.; Fritsch, L.; Young, R.; Morozova, N.; Fenouil, R.; Descostes, N.; Andrau, J.-C. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012, 19, 998. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. The three roles of RNA in protein synthesis. In Molecular Cell Biology, 4th ed.; WH Freeman: New York, NY, USA, 2000. [Google Scholar]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopper, A.K.; Pai, D.A.; Engelke, D.R. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010, 584, 310–317. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.-Y.; Ma, L.-M.; Chen, Y.-Q.; Qu, L.-H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef] [Green Version]
- Jöchl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Hüttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Chen, X.; Inukai, S.; Zhao, H.; Slack, F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 2011, 17, 1804–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babiarz, J.E.; Hsu, R.; Melton, C.; Thomas, M.; Ullian, E.M.; Blelloch, R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 2011, 17, 1489–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Bojanowski, K.; Ingber, D.E.; Panigrahy, D.; Pepper, M.S.; Montesano, R.; Shing, Y. New role for tRNA and its fragment purified from human urinary bladder carcinoma conditioned medium: Inhibition of endothelial cell growth. J. Cell. Biochem. 2000, 76, 109–117. [Google Scholar] [CrossRef]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 1–14. [Google Scholar] [CrossRef]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461–469. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, L.; Yu, X.; Ruan, Y.; Li, Z.; Guo, J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal. Transduct. Target. Ther. 2020, 5, 109. [Google Scholar] [CrossRef]
- Chen, C.J.; Heard, E. Small RNAs derived from structural non-coding RNAs. Methods 2013, 63, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Boskovic, A.; Bing, X.Y.; Kaymak, E.; Rando, O.J. Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev. 2020, 34, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Yu, J.; Zhou, P. Role of tRNA-derived fragments in cancer: Novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020, 10, 393–402. [Google Scholar]
- Papadimitriou, M.-A.; Avgeris, M.; Levis, P.; Papasotiriou, E.C.; Kotronopoulos, G.; Stravodimos, K.; Scorilas, A. tRNA-Derived Fragments (tRFs) in Bladder Cancer: Increased 5′-tRF-LysCTT Results in Disease Early Progression and Patients′ Poor Treatment Outcome. Cancers 2020, 12, 3661. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, H.; He, J.; Gao, W. Mechanisms of tRNA-derived fragments and tRNA halves in cancer treatment resistance. Biomark. Res. 2020, 8, 52. [Google Scholar] [CrossRef]
- Guo, Y.; Bosompem, A.; Mohan, S.; Erdogan, B.; Ye, F.; Vickers, K.C.; Sheng, Q.; Zhao, S.; Li, C.-I.; Su, P.-F.; et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genom. 2015, 16, 727. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.-S.; Lin, S.-Y.; Lieu, A.-S.; Wu, T.-L. Differential expression of human 5S snoRNA genes. Biochem. Biophys. Res. Commun. 2002, 299, 196–200. [Google Scholar] [CrossRef]
- Cao, P.; Yang, A.; Wang, R.; Xia, X.; Zhai, Y.; Li, Y.; Yang, F.; Cui, Y.; Xie, W.; Liu, Y. Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology 2018, 155, 542–556. [Google Scholar] [CrossRef]
- Langhendries, J.-L.; Nicolas, E.; Doumont, G.; Goldman, S.; Lafontaine, D.L. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget 2016, 7, 59519. [Google Scholar] [CrossRef] [Green Version]
- Pourebrahim, R.; Zhang, Y.; Liu, B.; Gao, R.; Xiong, S.; Lin, P.P.; McArthur, M.J.; Ostrowski, M.C.; Lozano, G. Integrative genome analysis of somatic p53 mutant osteosarcomas identifies Ets2-dependent regulation of small nucleolar RNAs by mutant p53 protein. Genes Dev. 2017, 31, 1847–1857. [Google Scholar] [CrossRef]
- LoRusso, P.M. Inhibition of the PI3K/AKT/mTOR pathway in solid tumors. J. Clin. Oncol. 2016, 34, 3803. [Google Scholar] [CrossRef]
- Wu, L.; Zheng, J.; Chen, P.; Liu, Q.; Yuan, Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed. Pharmacother. 2017, 90, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yang, D.; Luo, H.; Wu, S.; Dong, W.; Xiao, J.; Yuan, S.; Ni, A.; Zhang, K.-J.; Liu, X.-Y. SNORD126 promotes HCC and CRC cell growth by activating the PI3K–AKT pathway through FGFR2. J. Mol. Cell. Biol. 2017, 9, 243–255. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124. [Google Scholar] [CrossRef]
- Mannoor, K.; Shen, J.; Liao, J.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol. Cancer 2014, 13, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Zhang, J.; Ni, J.; Luo, J.; Wang, J.; Tang, L.; Zhang, L.; Wang, L.; Xu, J.; Su, B. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2015, 34, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorenoor, N.; Slaby, O. Small nucleolar RNAs functioning and potential roles in cancer. Tumor Biol. 2015, 36, 41–53. [Google Scholar] [CrossRef]
- Valleron, W.; Ysebaert, L.; Berquet, L.; Fataccioli, V.; Quelen, C.; Martin, A.; Parrens, M.; Lamant, L.; de Leval, L.; Gisselbrecht, C. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood J. Am. Soc. Hematol. 2012, 120, 3997–4005. [Google Scholar] [CrossRef]
- Yoshida, K.; Toden, S.; Weng, W.; Shigeyasu, K.; Miyoshi, J.; Turner, J.; Nagasaka, T.; Ma, Y.; Takayama, T.; Fujiwara, T. SNORA21—An oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine 2017, 22, 68–77. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, H.; Xie, Y.; Zhang, S.; Zhu, J.; Yin, G.; Shu, G.; Zhang, Y. Preliminary screening and identification of differentially expressed metastasis-related ncRNAs in ovarian cancer. Oncol. Lett. 2018, 15, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teittinen, K.J.; Laiho, A.; Uusimäki, A.; Pursiheimo, J.-P.; Gyenesei, A.; Lohi, O. Expression of small nucleolar RNAs in leukemic cells. Cell. Oncol. 2013, 36, 55–63. [Google Scholar] [CrossRef]
- Nallar, S.C.; Kalvakolanu, D.V. Regulation of snoRNAs in cancer: Close encounters with interferon. J. Interferon Cytokine Res. 2013, 33, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ruan, H.; Li, S.; Ye, Y.; Hong, W.; Gong, J.; Zhang, Z.; Jing, Y.; Zhang, X.; Diao, L. The genetic and pharmacogenomic landscape of snoRNAs in human cancer. Mol. Cancer 2020, 19, 108. [Google Scholar] [CrossRef]
- Mourksi, N.-E.-H.; Morin, C.; Fenouil, T.; Diaz, J.-J.; Marcel, V. snoRNAs offer novel insight and promising perspectives for lung cancer understanding and management. Cells 2020, 9, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.; Thompson, I.; Yuan, Z. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.-j.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.-Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.-T. Implication of snoRNA U50 in human breast cancer. J. Genet. Genom. 2009, 36, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Yang, J.; van Nues, R.; Watzinger, P.; Kötter, P.; Lafontaine, D.L.; Granneman, S.; Entian, K.-D. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017, 13, e1006804. [Google Scholar] [CrossRef]
- Dudnakova, T.; Dunn-Davies, H.; Peters, R.; Tollervey, D. Mapping targets for small nucleolar RNAs in yeast. Wellcome Open Res. 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Langhendries, J.L.; Watzinger, P.; Kötter, P.; Entian, K.D.; Lafontaine, D.L. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015, 43, 2242–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.-H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Lafontaine, D.L.J. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Newnam, G.P.; Liebman, S.W. The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: Genetic evidence in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 2517–2522. [Google Scholar] [CrossRef] [Green Version]
- Jorjani, H.; Kehr, S.; Jedlinski, D.J.; Gumienny, R.; Hertel, J.; Stadler, P.F.; Zavolan, M.; Gruber, A.R. An updated human snoRNAome. Nucleic Acids Res. 2016, 44, 5068–5082. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Kiss, T. Cooperative 2′-O-methylation of the wobble cytidine of human elongator tRNAMet (CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev. 2019, 33, 741–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, S.; Ivanov, P.; Hu, G.-F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Kiss, A.M.; Jády, B.E.; Bertrand, E.; Kiss, T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004, 24, 5797–5807. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Shen, M.; Gresch, P.J.; Ghamari-Langroudi, M.; Rabchevsky, A.G.; Emeson, R.B.; Stamm, S. Oligonucleotide-induced alternative splicing of serotonin 2C receptor reduces food intake. EMBO Mol. Med. 2016, 8, 878–894. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Stamm, S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef]
- Bratkovič, T.; Modic, M.; Ortega, G.C.; Drukker, M.; Rogelj, B. Neuronal differentiation induces SNORD115 expression and is accompanied by post-transcriptional changes of serotonin receptor 2c mRNA. Sci. Rep. 2018, 8, 5101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, C.M.; Chu, H.; Rueter, S.M.; Hutchinson, L.K.; Canton, H.; Sanders-Bush, E.; Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 1997, 387, 303–308. [Google Scholar] [CrossRef]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M.; Yamada, K.; Endo, A.; Barton, G.J.; Lamond, A.I. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012, 40, 3676–3688. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Shi, J.; Guo, Y.; Huang, W.; Huang, S.; Ming, S.; Wu, X.; Zhang, R.; Ding, J.; Zhao, W.; et al. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017, 45, 8647–8660. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Saraiya, A.A.; Wang, C.C. Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl. Trop. Dis. 2011, 5, e1338. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Pillman, K.A.; Anderson, M.L.; Lawrence, D.M.; Toubia, J.; Goodall, G.J.; Bracken, C.P. Assessing the gene regulatory properties of Argonaute-bound small RNAs of diverse genomic origin. Nucleic Acids Res. 2015, 43, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Macias, S.; Plass, M.; Stajuda, A.; Michlewski, G.; Eyras, E.; Cáceres, J.F. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012, 19, 760. [Google Scholar] [CrossRef] [Green Version]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Qian, S.B. Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 2014, 5, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mleczko, A.M.; Machtel, P.; Walkowiak, M.; Wasilewska, A.; Pietras, P.J.; Bąkowska-Żywicka, K. Levels of sdRNAs in cytoplasm and their association with ribosomes are dependent upon stress conditions but independent from snoRNA expression. Sci. Rep. 2019, 9, 18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plewka, P.; Szczesniak, M.W.; Stepien, A.; Zywicki, M.; Pacak, A.; Colombo, M.; Makalowska, I.; Ruepp, M.-D.; Raczynska, K.D. FUS controls the processing of snoRNAs into smaller RNA fragments that can regulate gene expression. bioRxiv 2019, 409250. [Google Scholar]
- Kulińska-Szweykowska, Z.; Kozlowski, P. Session 6. Non-coding RNA and Gene Expression. Acta Biochim. Pol. 2018, 65, 31–43. [Google Scholar]
- Sukhanova, M.V.; Singatulina, A.S.; Pastré, D.; Lavrik, O.I. Fused in Sarcoma (FUS) in DNA Repair: Tango with Poly(ADP-ribose) Polymerase 1 and Compartmentalisation of Damaged DNA. Int. J. Mol. Sci. 2020, 21, 7020. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Y.-C.; Mullane, P.; Ji, Y.J.; Liu, H.; He, L.; Arora, A.; Hwang, H.-Y.; Alessi, A.F.; Niaki, A.G.; et al. FUS Regulates Activity of MicroRNA-Mediated Gene Silencing. Mol. Cell 2018, 69, 787–801. [Google Scholar] [CrossRef] [Green Version]
- Falaleeva, M.; Surface, J.; Shen, M.; de la Grange, P.; Stamm, S. SNORD116 and SNORD115 change expression of multiple genes and modify each other’s activity. Gene 2015, 572, 266–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Hoogstrate, Y.; Kalsbeek, A.; Pigmans, B.; Vredenbregt-van den Berg, M.; Dits, N.; Nielsen, S.J.; Baker, A.; Visakorpi, T.; Bangma, C. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget 2015, 6, 17430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanov, G.A.; Filippova, J.A.; Komissarov, A.B.; Kuligina, E.V.; Richter, V.A.; Semenov, D.V. Regulatory role of small nucleolar RNAs in human diseases. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. https://doi.org/10.3390/ijms221910193
Wajahat M, Bracken CP, Orang A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. International Journal of Molecular Sciences. 2021; 22(19):10193. https://doi.org/10.3390/ijms221910193
Chicago/Turabian StyleWajahat, Maliha, Cameron Peter Bracken, and Ayla Orang. 2021. "Emerging Functions for snoRNAs and snoRNA-Derived Fragments" International Journal of Molecular Sciences 22, no. 19: 10193. https://doi.org/10.3390/ijms221910193
APA StyleWajahat, M., Bracken, C. P., & Orang, A. (2021). Emerging Functions for snoRNAs and snoRNA-Derived Fragments. International Journal of Molecular Sciences, 22(19), 10193. https://doi.org/10.3390/ijms221910193






