Biochemical Characterization of 13-Lipoxygenases of Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
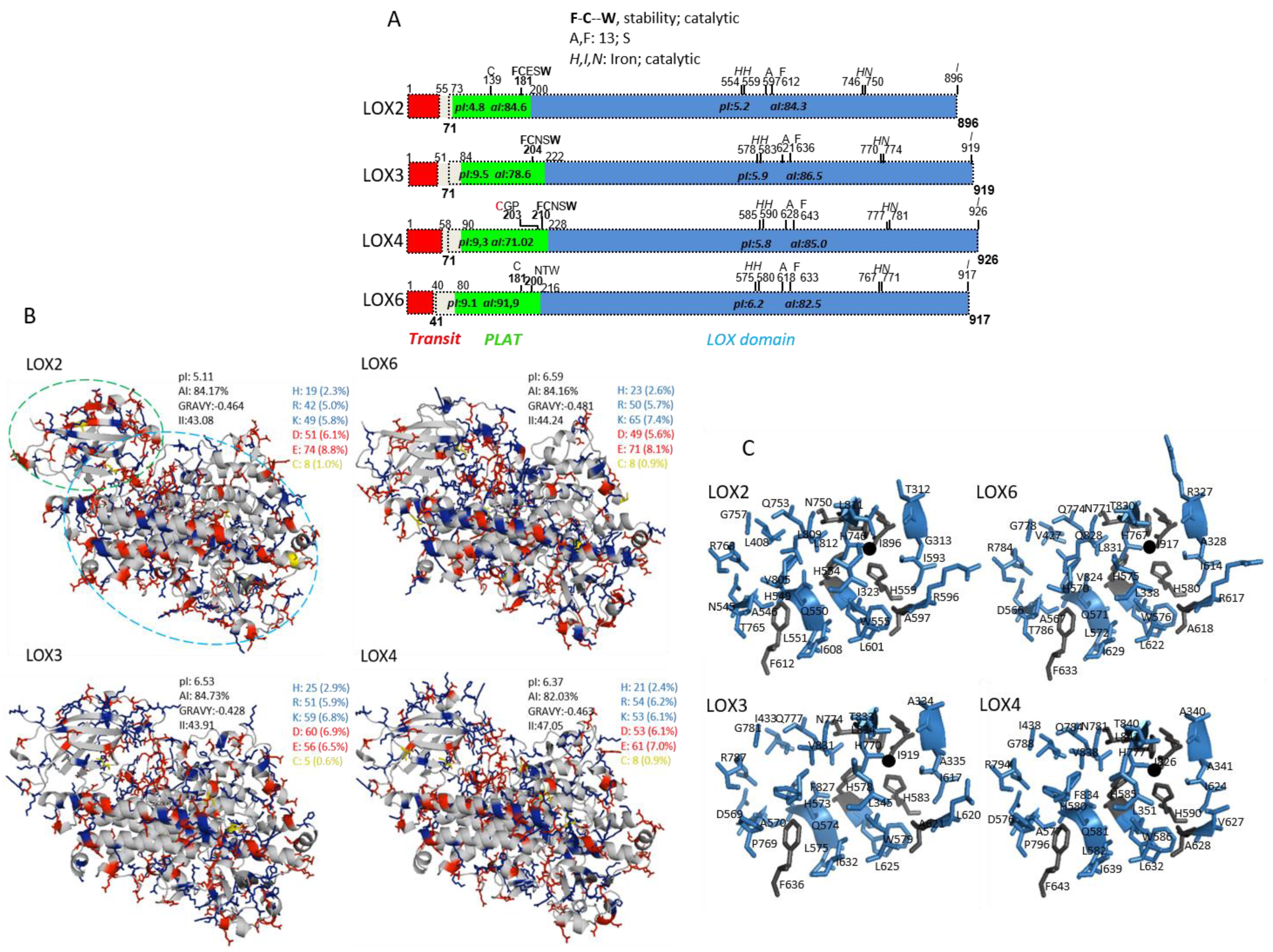
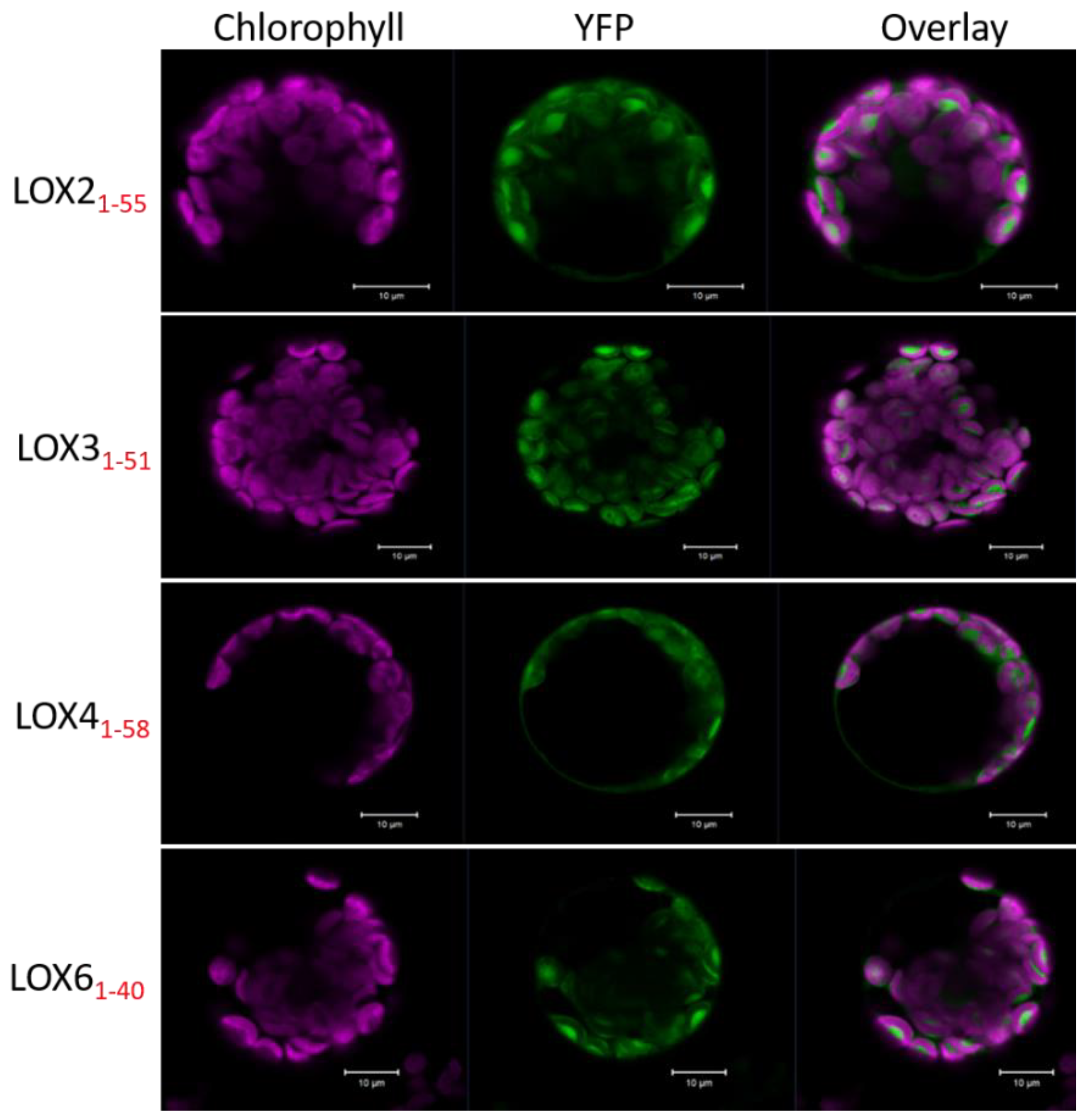
2.1. In Silico and Subcellular Targeting of Arabidopsis 13-Lipoxygenases
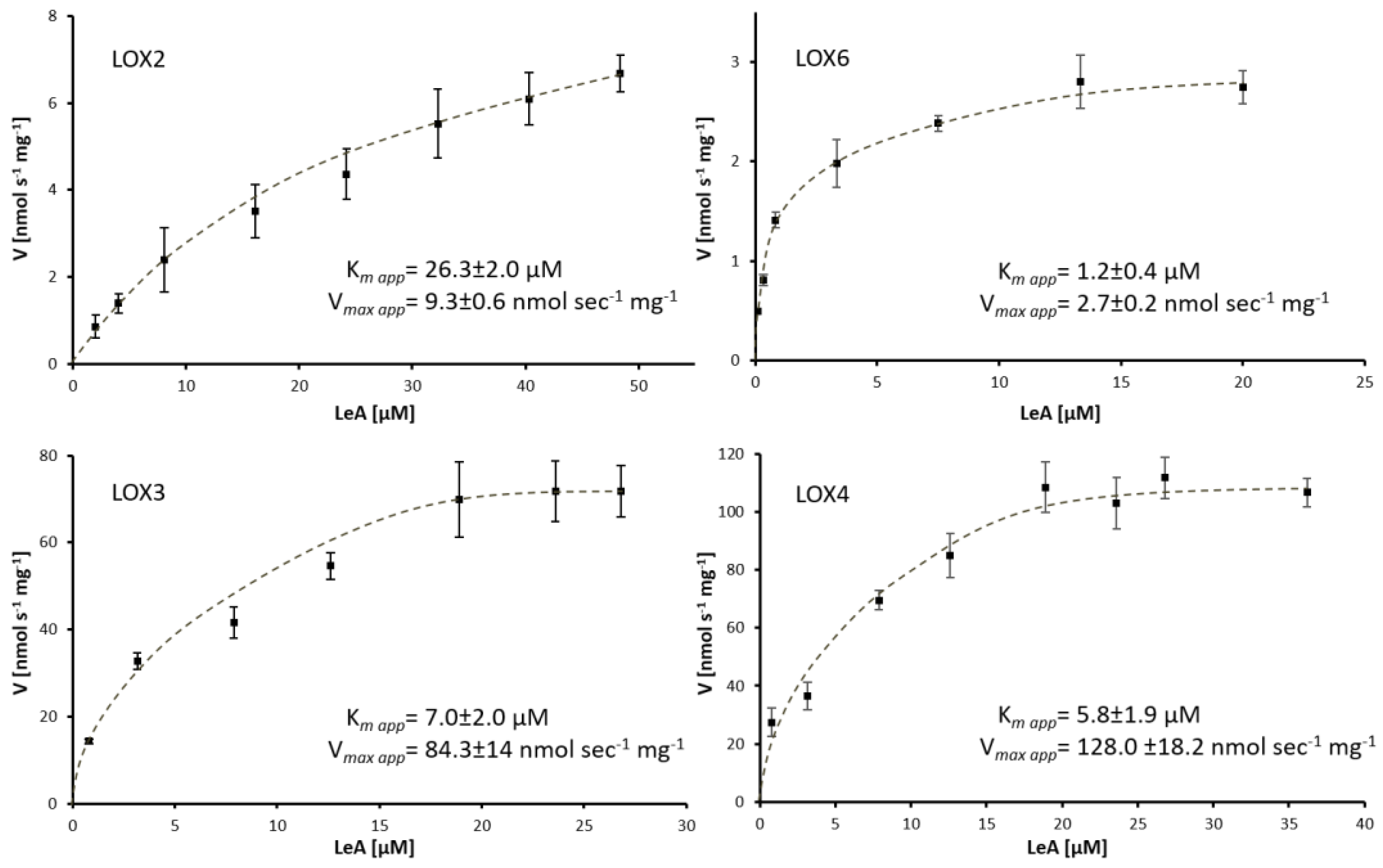
2.2. Purification and Enzymatic Characteristics of Arabidopsis 13-Lipoxygenases
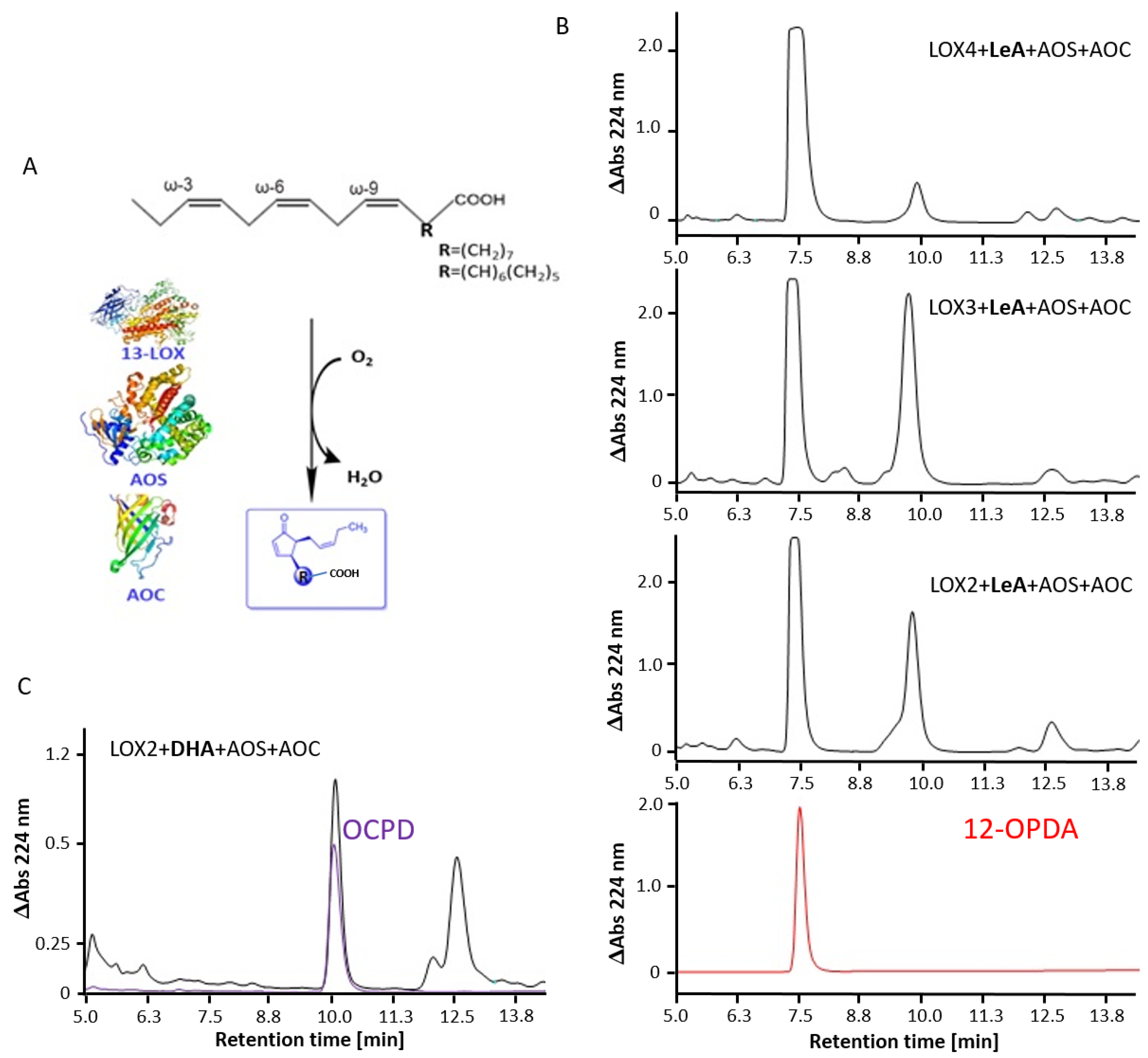
2.3. Arabidopsis 13-LOX Isoforms Mediate the Synthesis of 12-OPDA and OCPD
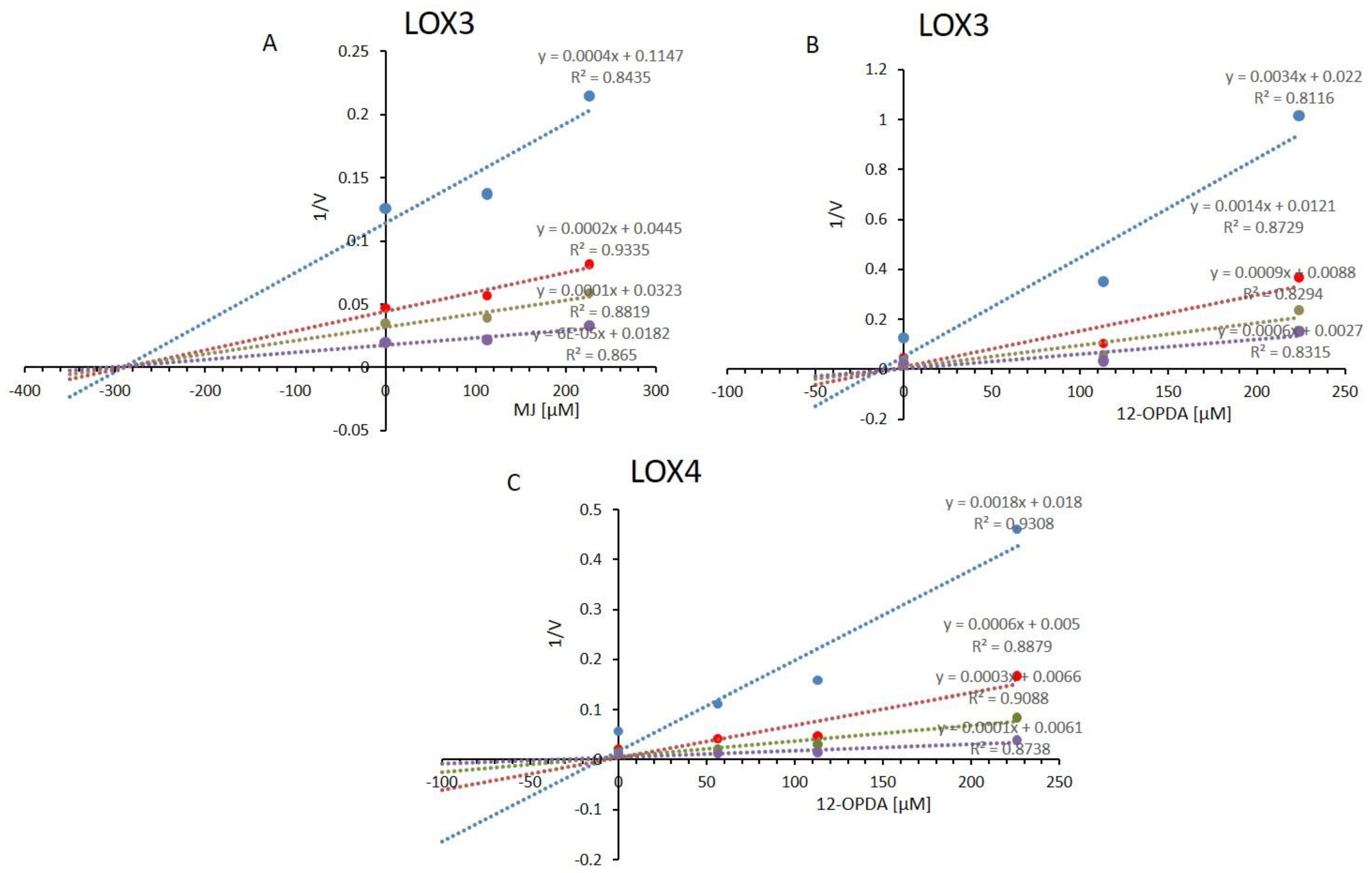
2.4. Effect of Metal Ions and LOX-Associated Phytohormones on Arabidopsis 13-LOX Activities
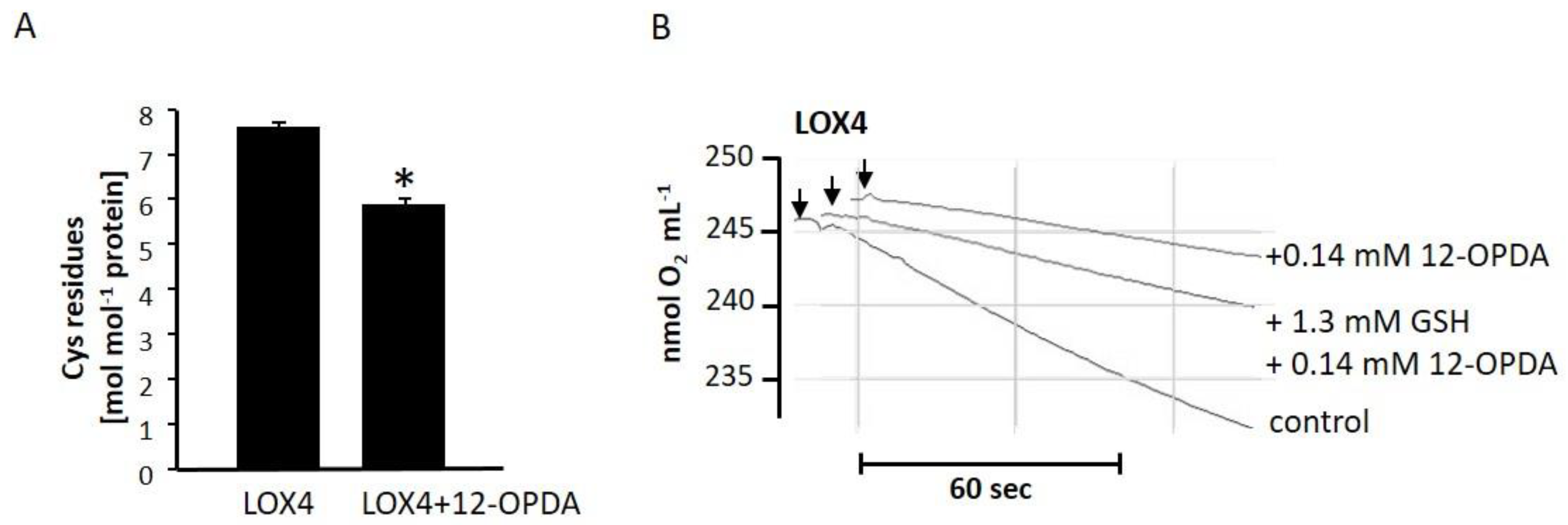
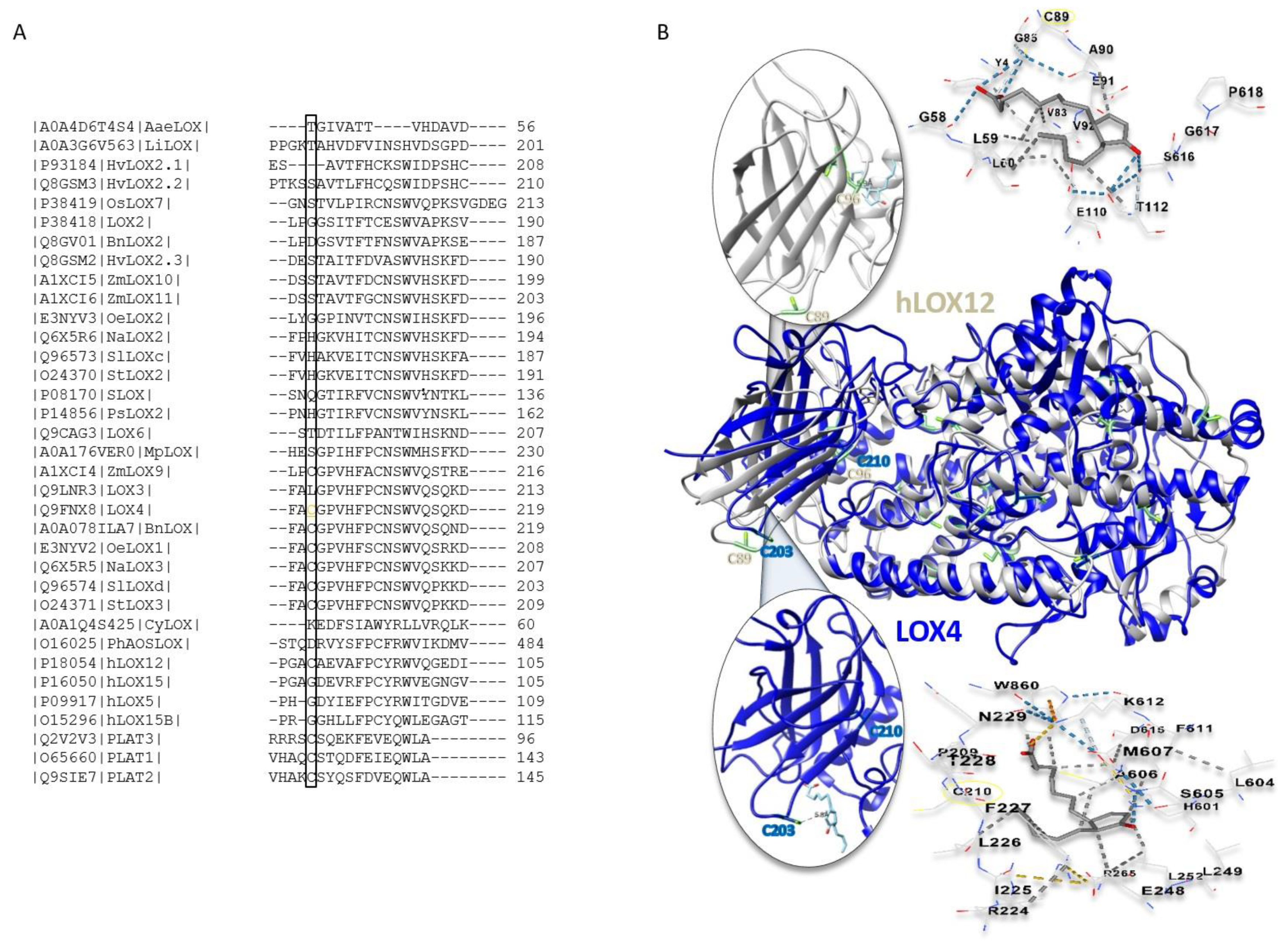
2.5. The Role of Cysteine Residues in Activity Regulation of AtLOX4
3. Discussion
3.1. Synthesis of Functional 13-LOX Isoforms
3.2. Subcellular Localization of 13-LOX Isoforms
3.3. Substrate Specificities and Evolutionary Relations
3.4. Differential Impact of Divalent Cations on 13-LOX Isoforms
3.5. Suicide Inhibition and Feedback Regulation
3.6. Cyclopentenone Prostaglandins as Potential LOX Inhibitors
4. Materials and Methods
4.1. Data processing, Databases and Computational Studies
4.2. Chemicals and Reagents
4.3. Subcellular Localization Studies
4.4. Cloning, Expression and Preparation of Lipoxygenase Wild-Type and Variant LOX2, LOX3, LOX4, LOX6 and LOX4 C203S
4.5. Lipoxygenase Activity
4.6. Effect of pH and Various Compounds on LOX Activities
4.7. Effect of Divalent Cations on LOX-Catalyzed LeA Dioxygenation
4.8. LOX-Mediated Synthesis of 12-OPDA, DHA-Peroxide, ARA-Peroxide and OCPD
4.9. Determination of Iron Content and Free Cysteines
4.10. Ex Vivo Evaluation of 12-OPDA as a LOX Inhibitor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horn, T.; Adel, S.; Schumann, R.; Sur, S.; Kakularam, K.R.; Polamarasetty, A.; Redanna, P.; Kuhn, H.; Heydeck, D. Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling. Prog. Lipid Res. 2015, 57, 13–39. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.D. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 2001, 294, 1871–1875. [Google Scholar] [CrossRef] [Green Version]
- Musiek, E.S.; Brooks, J.D.; Joo, M.; Brunoldi, E.; Porta, A.; Zanoni, G.; Vidary, G.; Blackwell, T.S.; Montine, T.J.; Milne, G.L.; et al. Electrophilic cyclopentenone neuroprostanes are anti-inflammatory mediators formed from the peroxidation of the ω-3 polyunsaturated fatty acid docosahexaenoic acid. J. Biol. Chem. 2008, 283, 19927–19935. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.M.; Zhu, Z.J.; Chen, J.J.; Yang, R.; Luo, Q.J.; Xu, J.L.; Shan, H.; Yan, X.J. A multifunctional lipoxygenase from Pyropia haitanensis—The cloned and functioned complex eukaryotic algae oxylipin pathway enzyme. Algal Res. 2015, 12, 316–327. [Google Scholar] [CrossRef]
- Djian, B.; Hornung, E.; Ischebeck, T.; Feussner, I. The green microalga Lobosphaera incisa harbours an arachidonate 15S-lipoxygenase. Plant Biol. 2019, 21, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhtarova, L.S.; Lantsova, N.V.; Khairutdinov, B.I.; Grechkin, A.N. Lipoxygenase pathway in model bryophytes: 12-oxo-9 (13), 15-phytodienoic acid is a predominant oxylipin in Physcomitrella patens. Phytochemistry 2020, 180, 112533. [Google Scholar] [CrossRef]
- Monte, I.; Ishida, S.; Zamarreño, A.M.; Hamberg, M.; Franco-Zorrilla, J.M.; García-Casado, G.; Gouhier-Darimont, C.; Reymond, P.; Takahashi, K.; García-Mina, J.M.; et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 2018, 14, 480–488. [Google Scholar] [CrossRef] [Green Version]
- Savadogo, E.H.; Shiomi, Y.; Yasuda, J.; Akino, T.; Yamaguchi, M.; Yoshida, H.; Umegawachi, T.; Tanaka, R.; Suong, D.N.A.; Miura, K.; et al. Gene expression of PLAT and ATS3 proteins increases plant resistance to insects. Planta 2021, 253, 1–16. [Google Scholar] [CrossRef]
- Ikei, K.N.; Yeung, J.; Apopa, P.L.; Ceja, J.; Vesci, J.; Holman, T.R.; Holinstat, M. Investigations of human platelet-type 12-lipoxygenase: Role of lipoxygenase products in platelet activation. J. Lipid Res. 2012, 53, 2546–2559. [Google Scholar] [CrossRef] [Green Version]
- Bonaventure, G.; Salas, J.J.; Pollard, M.R.; Ohlrogge, J.B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 2003, 15, 1020–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannenberg, G.; Martínez, M.; Hamberg, M.; Castresana, C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 2009, 44, 85. [Google Scholar] [CrossRef]
- Higashi, Y.; Okazaki, Y.; Takano, K.; Myouga, F.; Shinozaki, K.; Knoch, E.; Fukushima, A.; Saito, K. Heat inducible lipase1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α-linolenic acid in Arabidopsis leaves under heat stress. Plant Cell. 2018, 30, 1887–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutzner, L.; Goloshchapova, K.; Heydeck, D.; Stehling, S.; Kuhn, H.; Schebb, N.H. Mammalian ALOX15 orthologs exhibit pronounced dual positional specificity with docosahexaenoic acid. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Stelmach, B.A.; Müller, A.; Hennig, P.; Laudert, D.; Andert, L.; Weiler, E.W. Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry 1998, 47, 539–546. [Google Scholar] [CrossRef]
- Grebner, W.; Stingl, N.E.; Oenel, A.; Mueller, M.J.; Berger, S. Lipoxygenase 6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013, 161, 2159–2170. [Google Scholar] [CrossRef] [Green Version]
- Pollmann, S.; Springer, A.; Rustgi, S.; von Wettstein, D.; Kang, C.; Reinbothe, C.; Reinbothe, S. Substrate channeling in oxylipin biosynthesis through a protein complex in the plastid envelope of Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 1483–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, J.; Hamberg, M.; Miersch, O.; Parthier, B. Purification and characterization of allene oxide cyclase from dry corn seeds. Plant Physiol. 1997, 114, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2011, 75, 25–33. [Google Scholar] [CrossRef]
- Cao, S.; Chen, H.; Zhang, C.; Tang, Y.; Liu, J.; Qi, H. Heterologous expression and biochemical characterization of two lipoxygenases in oriental melon, Cucumis melo var. makuwa Makino. PLoS ONE 2016, 11, e0153801. [Google Scholar] [CrossRef]
- Wennman, A.; Oliw, E.H.; Karkehabadi, S.; Chen, Y. Crystal structure of manganese lipoxygenase of the rice blast fungus Magnaporthe oryzae. J. Biol. Chem. 2016, 291, 8130–8139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walther, M.; Wiesner, R.; Kuhn, H. Investigations into calcium-dependent membrane association of 15-lipoxygenase-1 mechanistic roles of surface-exposed hydrophobic amino acids and calcium. J. Biol. Chem. 2004, 279, 3717–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, J.B.; Brock, T.G. Structural organization of the regulatory domain of human 5-lipoxygenase. Curr. Protein Pept. Sci. 2005, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Eek, P.; Järving, R.; Järving, I.; Gilbert, N.C.; Newcomer, M.E.; Samel, N. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J. Biol. Chem. 2012, 287, 22377–22386. [Google Scholar] [CrossRef] [Green Version]
- Schaffrath, U.; Zabbai, F.; Dudler, R. Characterization of RCI-1, a chloroplastic rice lipoxygenase whose synthesis is induced by chemical plant resistance activators. Eur. J. Biochem. 2000, 267, 5935–5942. [Google Scholar] [CrossRef]
- Feussner, I.; Bachmann, A.; Höhne, M.; Kindl, H. All three acyl moieties of trilinolein are efficiently oxygenated by recombinant His-tagged lipid body lipoxygenase in vitro. FEBS Lett. 1998, 431, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Springer, A.; Kang, C.; Rustgi, S.; von Wettstein, D.; Reinbothe, C.; Pollmann, S.; Reinbothe, S. Programmed chloroplast destruction during leaf senescence involves 13-lipoxygenase (13-LOX). Proc. Natl. Acad. Sci. USA 2016, 113, 3383–3388. [Google Scholar] [CrossRef] [Green Version]
- Kühn, H.; Barnett, J.; Grunberger, D.; Baecker, P.; Chow, J.; Nguyen, B.; Bursztyn-Pettegrew, H.; Chan, H.; Sigal, E. Overexpres-sion, purification and characterization of human recombinant 15-lipoxygenase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1993, 1169, 80–89. [Google Scholar] [CrossRef]
- Goloshchapova, K.; Stehling, S.; Heydeck, D.; Blum, M.; Kuhn, H. Functional characterization of a novel arachidonic acid 12S-lipoxygenase in the halotolerant bacterium Myxococcus fulvus exhibiting com-plex social living patterns. MicrobiologyOpen 2019, 8, e00775. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Hamberg, M. Specificity of two lipoxygenases from rice: Unusual regiospecificity of a lipoxygenase isoenzyme. Lipids 1996, 31, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, J.; Wasternack, C.; Hamberg, M. On the specificity of allene oxide cyclase. Lipids 1999, 34, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Maynard, D.; Müller, S.M.; Hahmeier, M.; Löwe, J.; Feussner, I.; Gröger, H.; Viehhauser, A.; Dietz, K.J. One-pot synthesis of bioactive cyclopentenones from α-linolenic acid and docosahexaenoic acid. Bioorg. Med. Chem. 2018, 7, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, S.; Uchibori, A.; Takagi, H.; Maki, R.; Ohnishi, M. Light-induced increase in free Mg2+ concentration in spinach chloroplasts: Measurement of free Mg2+ by using a fluorescent probe and necessity of stromal alkalinization. Arch. Biochem. Biophys. 2003, 412, 126–132. [Google Scholar] [CrossRef]
- Tatulian, S.A.; Steczko, J.; Minor, W. Uncovering a calcium-regulated membrane-binding mechanism for soybean lipoxygenase-1. Biochemistry 1998, 37, 15481–15490. [Google Scholar] [CrossRef]
- Hammarberg, T.; Provost, P.; Persson, B.; Rådmark, O. The N-terminal domain of 5-lipoxygenase binds calcium and mediates calcium stimulation of enzyme activity. J. Biol. Chem. 2000, 275, 38787–38793. [Google Scholar] [CrossRef] [Green Version]
- Mueller, M.J.; Berger, S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry 2009, 70, 1511–1521. [Google Scholar] [CrossRef]
- Kuninori, T.; Nishiyama, J.; Shirakawa, M.; Shimoyama, A. Inhibition of soybean lipoxygenase-1 by n-alcohols and n-alkylthiols. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1992, 1125, 49–55. [Google Scholar] [CrossRef]
- Berger, S.; Weichert, H.; Porzel, A.; Wasternack, C.; Kühn, H.; Feussner, I. Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2001, 1533, 266–276. [Google Scholar] [CrossRef]
- Sanchez-Arcos, C.; Reichelt, M.; Gershenzon, J.; Kunert, G. Modulation of legume defense signaling pathways by native and non-native pea aphid clones. Front. Plant Sci. 2016, 7, 1872. [Google Scholar] [CrossRef] [Green Version]
- Cohen, B.S.; Grossman, S.; Pinsky, A. Chlorophyll inhibition of lipoxygenase in growing pea plants. J. Agric. Food Chem. 1984, 32, 516–519. [Google Scholar] [CrossRef]
- Maucher, I.V.; Ruehl, M.; Kretschmer, S.B.; Hofmann, B.; Kühn, B.; Fettel, J.; Vogel, A.; Flügel, K.; Manolikakes, G.; Hellmuth, N.; et al. Michael acceptor containing drugs are a novel class of 5-lipoxygenase inhibitor targeting the surface cysteines C416 and C418. Biochem. Pharmacol. 2017, 125, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.C.; Gerstmeier, J.; Schexnaydre, E.E.; Börner, F.; Garscha, U.; Neau, D.B.; Werz, O.; Newcomer, M.E. Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products. Nat. Chem. Biol. 2020, 16, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Fan, Y.; Gu, T.; Heydeck, D.; Stehling, S.; Ivanov, I.; Yao, Y.G.; Kuhn, H. The lipoxygenase pathway of Tupaia belangeri representing Scandentia. Genomic multiplicity and functional characterization of the ALOX15 orthologs in the tree shrew. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158550. [Google Scholar] [CrossRef] [PubMed]
- König, S.; Romp, E.; Krauth, V.; Rühl, M.; Dörfer, M.; Liening, S.; Hoffman, B.; Häfner, A.K.; Steinhilber, D.; Karas, M.; et al. Melleolides from honey mushroom inhibit 5-lipoxygenase via CYS159. Cell Chem. Biol. 2019, 26, 60–70. [Google Scholar] [CrossRef]
- Marvian-Hosseini, Z.; Asoodeh, A. Biochemical characterization of purified lipoxygenase from sesame (Sesamum indicum). Int. J. Food Prop. 2017, 20, S948–S958. [Google Scholar] [CrossRef]
- Macri, F.; Braidot, E.; Petrussa, E.; Vianello, A. Lipoxygenase activity associated to isolated soy-bean plasma membranes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1994, 1215, 109–114. [Google Scholar] [CrossRef]
- Yamamoto, S. Mammalian lipoxygenases: Molecular structures and functions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1992, 1128, 117–131. [Google Scholar] [CrossRef]
- Peltier, J.B.; Ytterberg, A.J.; Sun, Q.; van Wijk, K.J. New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 2004, 279, 49367–49383. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, J.; Trigueros, M.; Rojas-Triana, M.; Fernández, M.; Albar, J.P.; Bustos, R.; Paz-Ares, J.; Rubio, V. Proteomics identifies ubiquitin–proteasome targets and new roles for chromatin-remodeling in the Arabidopsis response to phosphate starvation. J. Proteom. 2013, 94, 1–22. [Google Scholar] [CrossRef]
- Royo, J.; Vancanneyt, G.; Pérez, A.G.; Sanz, C.; Störmann, K.; Rosahl, S.; Sánchez-Serrano, J.J. Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J. Biol. Chem. 1996, 271, 21012–21019. [Google Scholar] [CrossRef] [Green Version]
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013, 197, 566–575. [Google Scholar] [CrossRef]
- Friedman, R. Structural and computational insights into the versatility of cadmium binding to proteins. Dalton Trans. 2014, 43, 2878–2887. [Google Scholar] [CrossRef]
- Ritter, A.; Goulitquer, S.; Salaün, J.P.; Tonon, T.; Correa, J.A.; Potin, P. Copper stress induces biosynthesis of octadecanoid and eicosanoid oxygenated derivatives in the brown algal kelp Laminaria digitata. New Phytol. 2008, 180, 809–821. [Google Scholar] [CrossRef]
- Yan, J.; Chia, J.C.; Sheng, H.; Jung, H.I.; Zavodna, T.O.; Zhang, L.; Huang, R.; Jiao, C.; Craft, E.J.; Fei, Z.; et al. Arabidopsis pollen fertility requires the transcription factors CITF1 and SPL7 that regulate copper delivery to anthers and jasmonic acid synthesis. Plant Cell. 2017, 29, 3012–3029. [Google Scholar] [CrossRef] [Green Version]
- Lei, G.J.; Sun, L.; Sun, Y.; Zhu, X.F.; Li, G.X.; Zheng, S.J. Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 2020, 62, 218–227. [Google Scholar] [CrossRef]
- Montillet, J.L.; Cacas, J.L.; Garnier, L.; Montané, M.H.; Douki, T.; Bessoule, J.J.; Kowalczyck, L.P.; Maciejewska, U.; Agnel, J.P.; Vial, A.; et al. The upstream oxylipin profile of Arabidopsis thaliana: A tool to scan for oxidative stresses. Plant J. 2004, 40, 439–451. [Google Scholar] [CrossRef]
- Wu, H. Affecting the activity of soybean lipoxygenase-1. J. Mol. Graph. Model 1996, 14, 331–337. [Google Scholar] [CrossRef]
- Conrad, D.J. The arachidonate 12/15 lipoxygenases. Clin. Rev. Allergy Immunol. 1999, 17, 71–89. [Google Scholar] [CrossRef]
- Chauvin, A.; Lenglet, A.; Wolfender, J.L.; Farmer, E. Paired hierarchical organization of 13-lipoxygenases in Arabidopsis. Plants 2016, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Egan, R.W.; Gale, P.H. Inhibition of mammalian 5-lipoxygenase by aromatic disulfides. J. Biol. Chem. 1985, 260, 11554–11559. [Google Scholar] [CrossRef]
- Flatman, S.; Hurst, J.S.; McDonald-Gibson, R.G.; Jonas, G.E.; Slater, T.F. Biochemical studies on a 12-lipoxygenase in human uterine cervix. Biochim. Biophys. Acta Gen. Subj. 1986, 883, 7–14. [Google Scholar] [CrossRef]
- Hardy, D.J.; Gallegos, M.V.; Gaunt, J.K. Extracts of Pisum sativum metabolise phospholipid via a lipoxygenase-like mechanism. Phytochemistry 1991, 30, 2889–2894. [Google Scholar] [CrossRef]
- Bell, E.; Mullet, J.E. Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 1993, 103, 1133–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inzé, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mphahlele, M.J.; Gildenhuys, S.; Agbo, E.N. In vitro evaluation and docking studies of 5-oxo-5H-furo [3, 2-g] chromene-6-carbaldehyde derivatives as potential anti-Alzheimer’s agents. Int. J. Mol. Sci. 2019, 20, 5451. [Google Scholar] [CrossRef] [Green Version]
- Mogul, R.; Johansen, E.; Holman, T.R. Oleyl sulfate reveals allosteric inhibition of soybean lipoxygenase-1 and human 15-lipoxygenase. Biochemistry 2000, 39, 4801–4807. [Google Scholar] [CrossRef]
- Löwe, J.; Dietz, K.J.; Gröger, H. From a biosynthetic pathway toward a biocatalytic process and chemocatalytic modifications: Three-step enzymatic cascade to the plant metabolite cis-(+)-12-OPDA and metathesis-derived products. Adv. Sci. 2020, 7, 1902973. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Grimm, M.; Dai, W.-T.; Hou, M.-C.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2019, 41, 138–144. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Lu, W.; Zhao, X.; Xu, Z.; Dong, N.; Zou, S.; Shen, X.; Huang, J. Development of a new colorimetric assay for lipoxygenase activity. Anal. Biochem. 2013, 441, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Seidel, T.; Kluge, C.; Hanitzsch, M.; Roß, J.; Sauer, M.; Dietz, K.J.; Golldack, D. Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J. Biotechnol. 2004, 112, 165–175. [Google Scholar] [CrossRef]
- Vick, B.A. A spectrophotometric assay for hydroperoxide lyase. Lipids 1991, 26, 315–320. [Google Scholar] [CrossRef]
- Ferreira, C.M.; Pinto, I.S.; Soares, E.V.; Soares, H.M. (Un) suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions—A review. RSC Adv. 2015, 5, 30989–31003. [Google Scholar] [CrossRef] [Green Version]
- Fish, W.W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988, 158, 357–364. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Michelet, L.; Massot, V.; Trost, P.; Lemaire, S.D. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J. Biol. Chem. 2008, 283, 8868–8876. [Google Scholar] [CrossRef] [Green Version]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [Green Version]

| PUFA | Relative Activity (%) | |||
|---|---|---|---|---|
| LOX2 | LOX3 | LOX4 | LOX6 | |
| LeA | 100 | 100 | 100 | 100 |
| LA | 79 ± 7 | 12 ± 1 | 11 ± 1 | 17 ± 1 |
| ARA | 50 ± 4 | 5 ± 1 | 4 ± 1 | 8 ± 3 |
| DHA | 56 ± 3 | 7 ± 1 | 8 ± 1 | 31 ± 3 |
| Metal Salts | Relative Activity (%) | |||
|---|---|---|---|---|
| LOX2 | LOX3 | LOX4 | LOX6 | |
| CaCl2 (1 mM) | 690 ± 60% * | 117 ± 10% * | 100 ± 4% | 121 ± 15% * |
| MgCl2 (1 mM) | 170 ± 18% * | 135 ± 4% * | 106 ± 3% * | 142 ± 15% * |
| CdSO4 (1 mM) | 220 ± 18% * | no activity | no activity | no activity |
| CuCl2 (100 µM) | 50 ± 20% * | no activity | no activity | no activity |
| Polarography | Relative Activity (%) | |||
| LOX2 | LOX3 | LOX4 | LOX6 | |
| NEM | 84 ± 4% * | 51 ± 6% * | 11 ± 3% * | 47 ± 15% * |
| 12-OPDA | 64 ± 4% * | 60 ± 5% * | 21 ± 5% * | 50 ± 10% * |
| MJ | 87 ± 10% * | 85 ± 5% * | 95 ± 3% | 94 ± 6% |
| TA | 95 ± 8% | 100 ± 5% | 100 ± 9% | 101 ± 12% |
| SA | 96 ± 7% | 101 ± 5% | 101 ± 4% | 103 ± 5% |
| Photometry | Relative activity (%) | |||
| LOX2 | LOX3 | LOX4 | LOX6 | |
| NEM | 35 ± 10% * | 29 ± 10% * | 3 ± 2% * | 13 ± 2% * |
| 12-OPDA | 49 ± 10% * | 35 ± 5% * | 6 ± 4% * | 59 ± 10% * |
| MJ | 62 ± 10% * | 60 ± 4% * | 99 ± 7% | 86 ± 6% * |
| TA | 105 ± 5% | 103 ± 4% | 106 ± 4% | 100 ± 9% |
| SA | 100 ± 8% | 106 ± 5% | 103 ± 8% | 100 ± 9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maynard, D.; Chibani, K.; Schmidtpott, S.; Seidel, T.; Spross, J.; Viehhauser, A.; Dietz, K.-J. Biochemical Characterization of 13-Lipoxygenases of Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 10237. https://doi.org/10.3390/ijms221910237
Maynard D, Chibani K, Schmidtpott S, Seidel T, Spross J, Viehhauser A, Dietz K-J. Biochemical Characterization of 13-Lipoxygenases of Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(19):10237. https://doi.org/10.3390/ijms221910237
Chicago/Turabian StyleMaynard, Daniel, Kamel Chibani, Sonja Schmidtpott, Thorsten Seidel, Jens Spross, Andrea Viehhauser, and Karl-Josef Dietz. 2021. "Biochemical Characterization of 13-Lipoxygenases of Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 19: 10237. https://doi.org/10.3390/ijms221910237
APA StyleMaynard, D., Chibani, K., Schmidtpott, S., Seidel, T., Spross, J., Viehhauser, A., & Dietz, K.-J. (2021). Biochemical Characterization of 13-Lipoxygenases of Arabidopsis thaliana. International Journal of Molecular Sciences, 22(19), 10237. https://doi.org/10.3390/ijms221910237







