RHAMM Is a Multifunctional Protein That Regulates Cancer Progression
Abstract
:1. Introduction
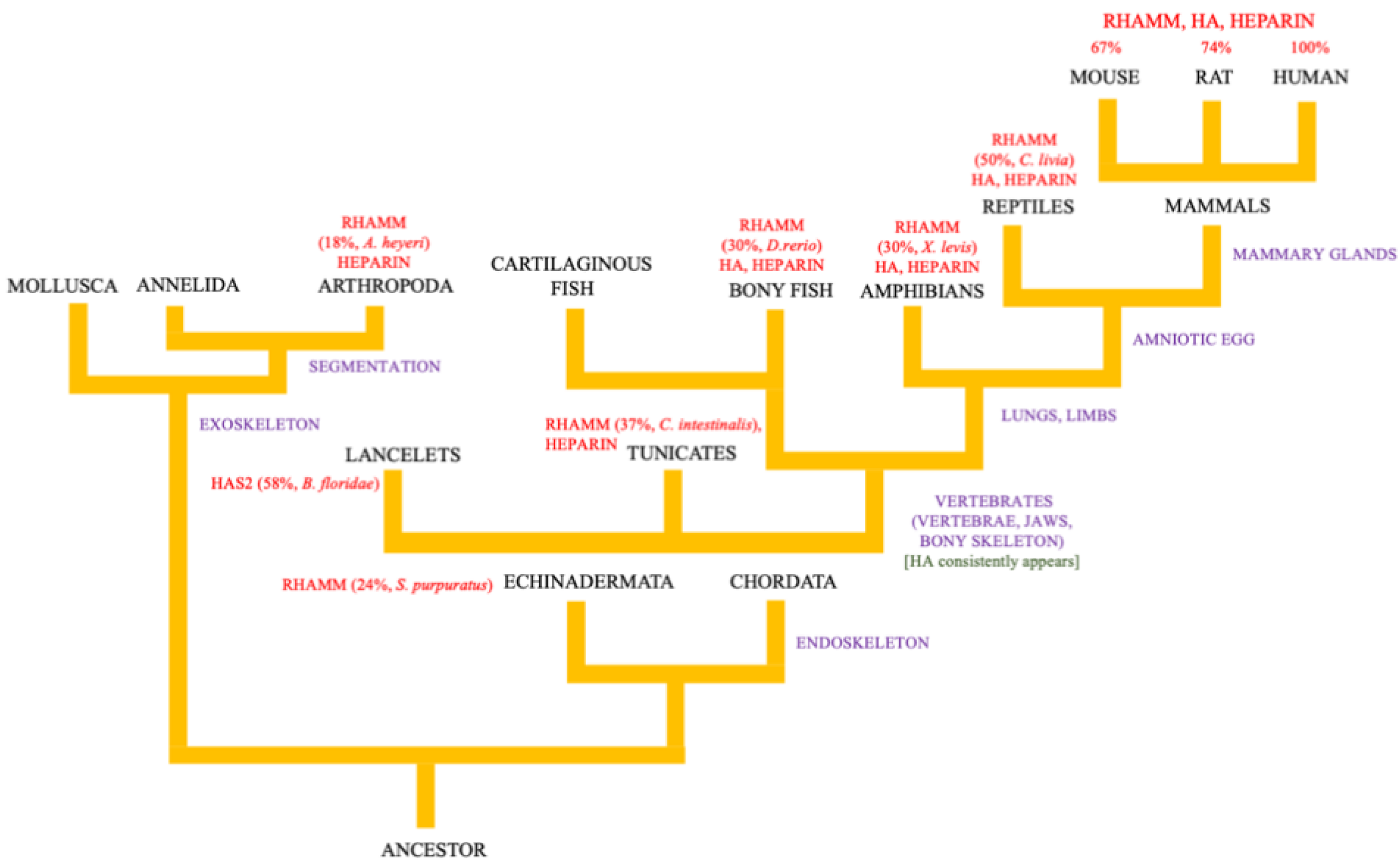
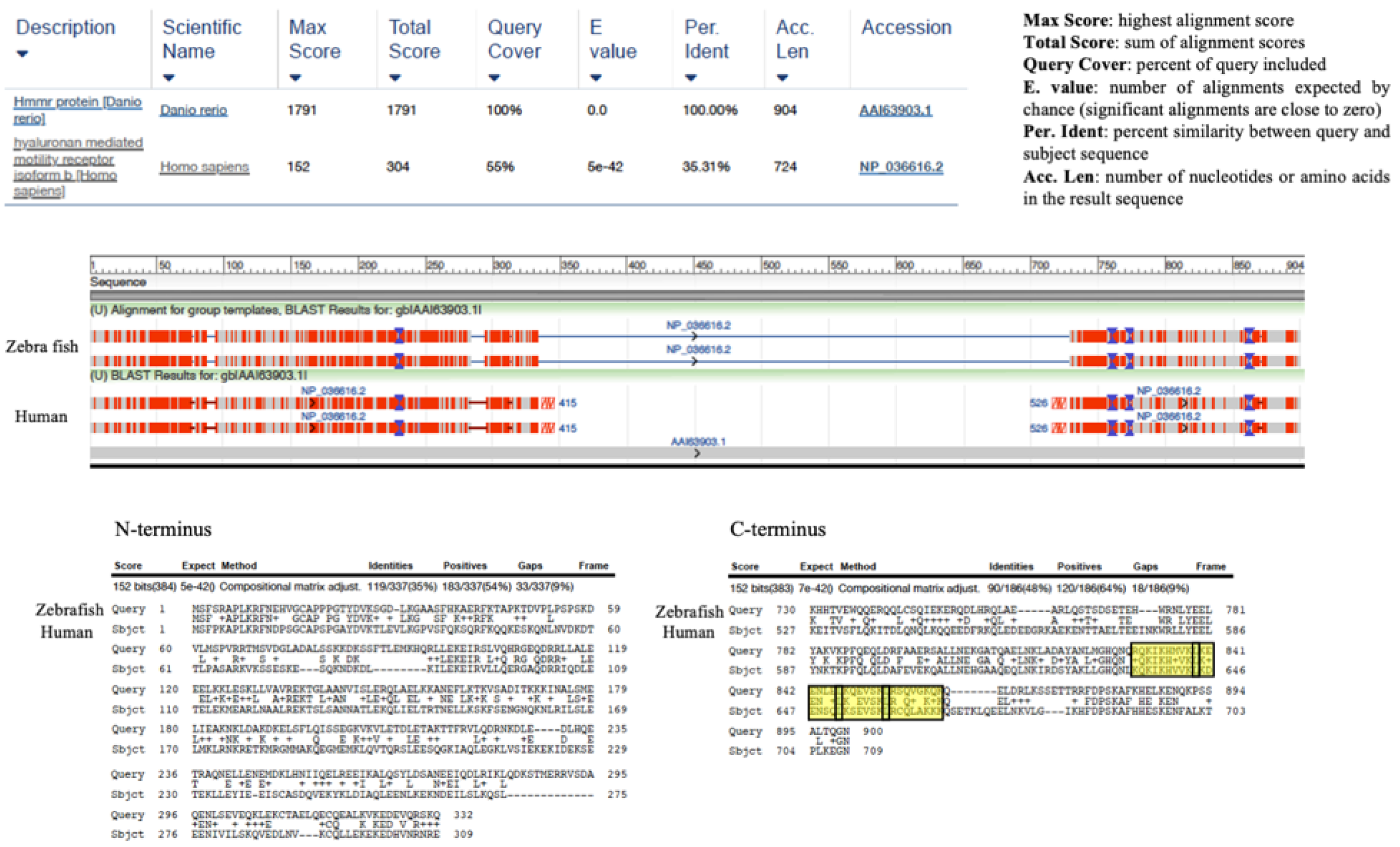
2. Evolution of the Hyaluronome
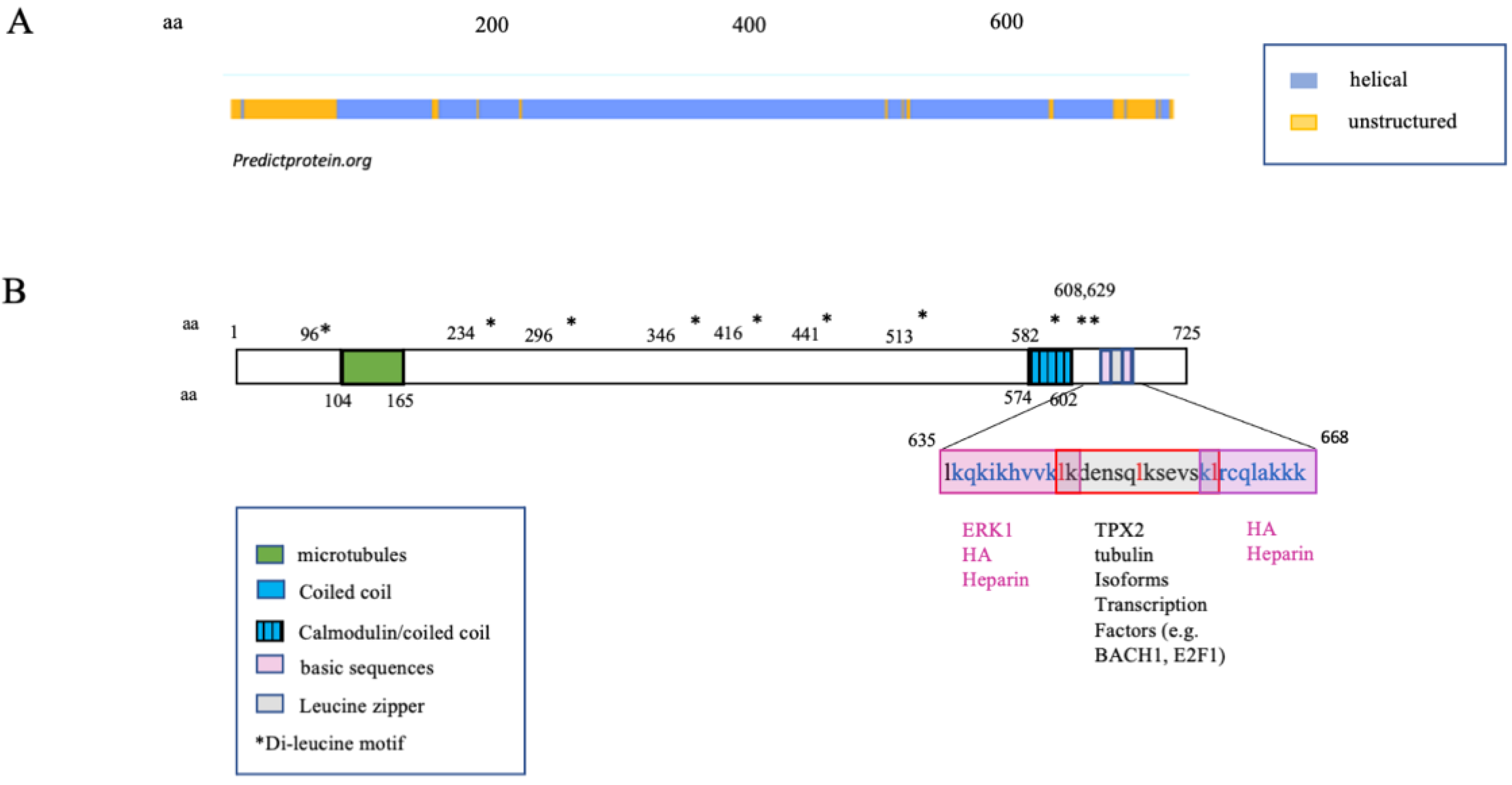
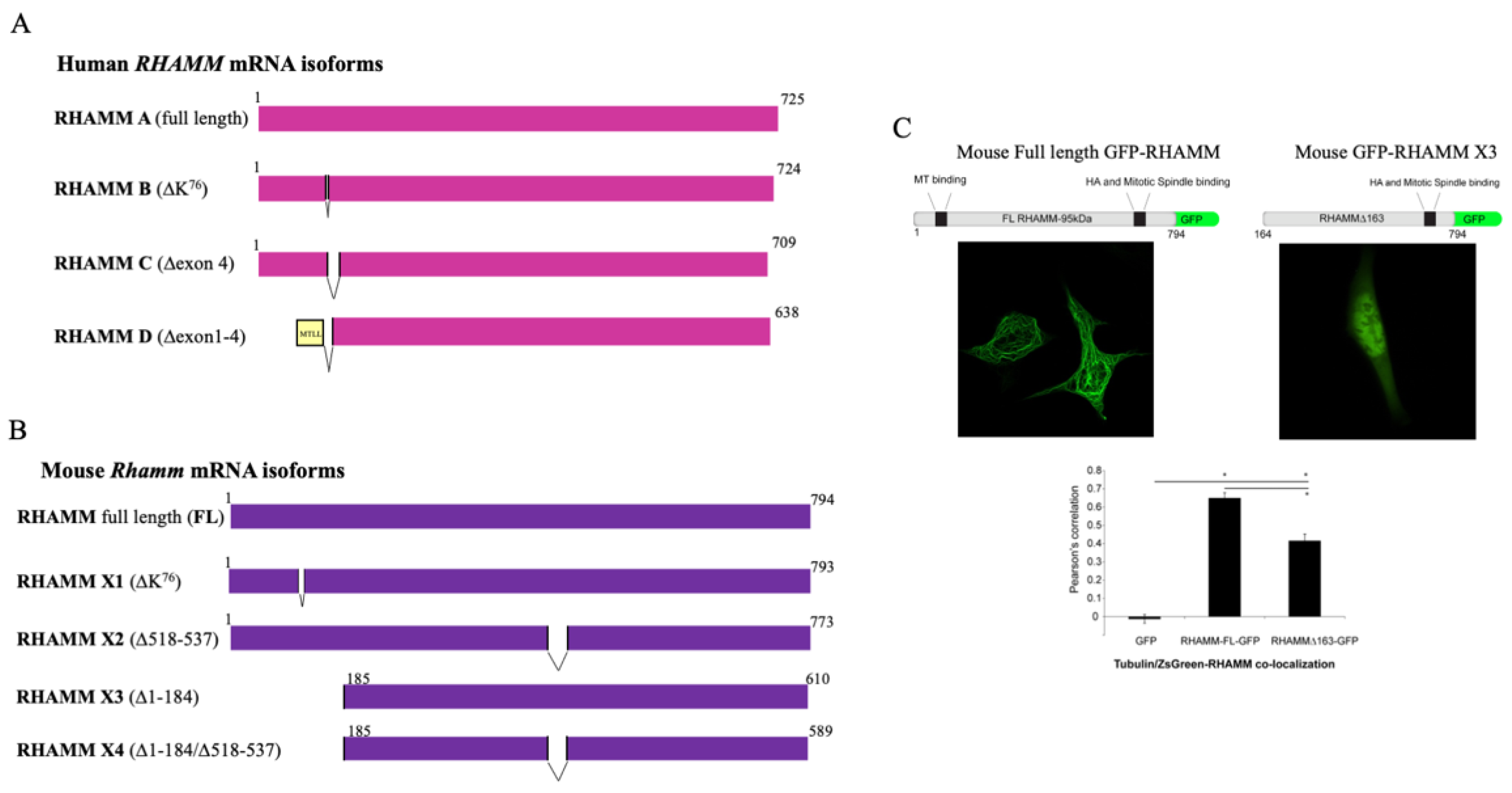
3. RHAMM Isoform Structure and Function
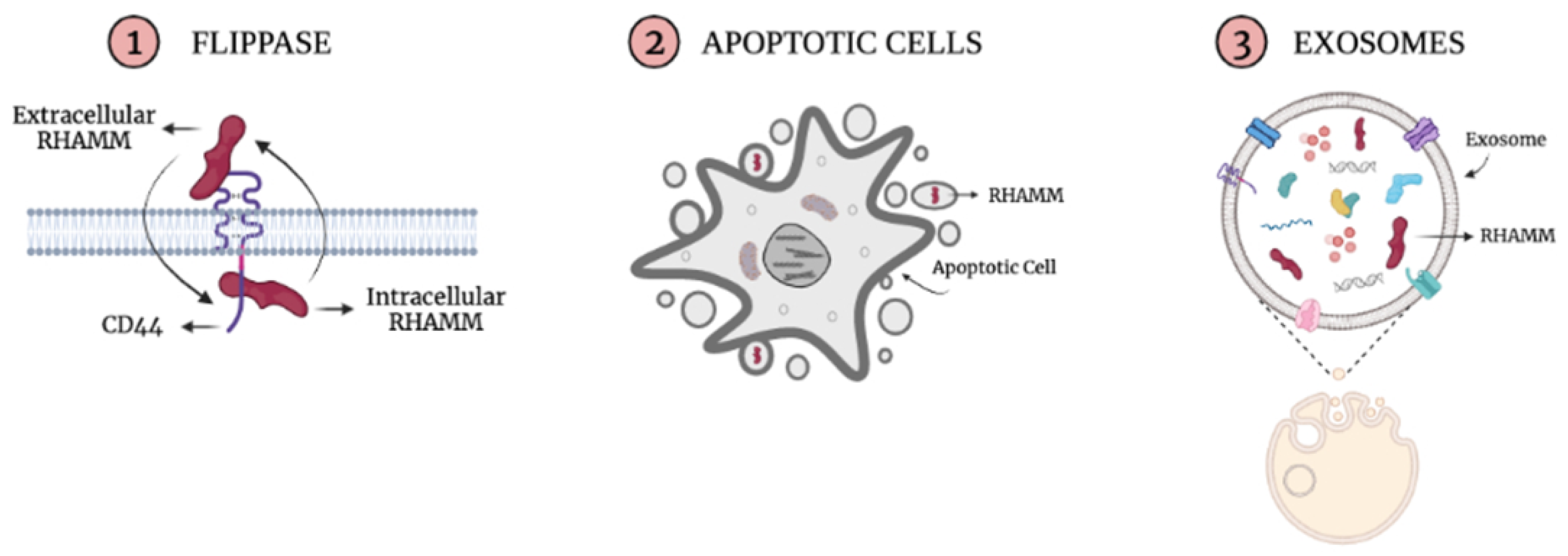
4. RHAMM Multi-Functions and Domains
5. RHAMM Multi-Functions in Cancer
6. Therapeutic Strategies for Targeting RHAMM
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beadle, G.W.; Tatum, E.L. Genetic Control of Biochemical Reactions in Neurospora. Proc. Natl. Acad. Sci. USA 1941, 27, 499–506. [Google Scholar] [CrossRef] [Green Version]
- Espinosa-Cantu, A.; Cruz-Bonilla, E.; Noda-Garcia, L.; DeLuna, A. Multiple Forms of Multifunctional Proteins in Health and Disease. Front. Cell Dev. Biol. 2020, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Radisky, D.C.; Stallings-Mann, M.; Hirai, Y.; Bissell, M.J. Single proteins might have dual but related functions in intracellular and extracellular microenvironments. Nat. Rev. Mol. Cell Biol. 2009, 10, 228–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.; Wang, D.; Chen, X.; Tang, L.H.; Verma, A.; Chen, Z.; Kim, B.J.; Selesner, L.; Robzyk, K.; Zhang, G.; et al. Function and clinical relevance of RHAMM isoforms in pancreatic tumor progression. Mol. Cancer 2019, 18, 92. [Google Scholar] [CrossRef] [Green Version]
- Hall, C.L.; Yang, B.; Yang, X.; Zhang, S.; Turley, M.; Samuel, S.; Lange, L.A.; Wang, C.; Curpen, G.D.; Savani, R.C.; et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell 1995, 82, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, C.A.; McCarthy, J.; Turley, E. Cell-surface and mitotic-spindle RHAMM: Moonlighting or dual oncogenic functions? J. Cell Sci. 2008, 121 Pt 7, 925–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Feng, J.; Chen, X.; Wang, D.; Wong, M.; Zhang, G.; Na, J.; Zhang, T.; Chen, Z.; Chen, Y.T.; et al. High levels of truncated RHAMM cooperate with dysfunctional p53 to accelerate the progression of pancreatic cancer. Cancer Lett. 2021, 514, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Liu, M.; Cousteils, K.; Telmer, P.; Alam, K.; Ma, J.; Mendina, L.; McCarthy, J.B.; Morris, V.L.; Turley, E.A. Cell-specific expression of the transcriptional regulator RHAMM provides a timing mechanism that controls appropriate wound re-epithelialization. J. Biol. Chem. 2020, 295, 5427–5448. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, D.; Shen, W.; Sun, W.; Gao, R.; Jiang, P.; Wang, L.; Liu, Y.; Chen, Y.; Zhou, W.; et al. RHAMM inhibits cell migration via the AKT/GSK3beta/Snail axis in luminal A subtype breast cancer. Anat. Rec. 2020, 303, 2344–2356. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Hamilton, S.R.; Nakrieko, K.A.; Kooshesh, F.; Walton, P.; McCarthy, J.B.; Bissell, M.; Turley, E.A. Rhamm−/− fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J. Cell Biol. 2006, 175, 1017–1028. [Google Scholar] [CrossRef] [Green Version]
- Hardwick, C.; Hoare, K.; Owens, R.; Hohn, H.P.; Hook, M.; Moore, D.; Cripps, V.; Austen, L.; Nance, D.M.; Turley, E.A. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J. Cell Biol. 1992, 117, 1343–1350. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Tolg, C.; Turley, E. Dissecting the Dual Nature of Hyaluronan in the Tumor Microenvironment. Front. Immunol. 2019, 10, 947. [Google Scholar] [CrossRef]
- Garantziotis, S.; Brezina, M.; Castelnuovo, P.; Drago, L. The role of hyaluronan in the pathobiology and treatment of respiratory disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 310, L785–L795. [Google Scholar] [CrossRef] [Green Version]
- Garantziotis, S.; Savani, R.C. Hyaluronan biology: A complex balancing act of structure, function, location and context. Matrix Biol. 2019, 78–79, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular size-dependent signaling and biological functions in inflammation and cancer. FEBS J. 2019, 286, 2883–2908. [Google Scholar] [CrossRef]
- Nikitovic, D.; Tzardi, M.; Berdiaki, A.; Tsatsakis, A.; Tzanakakis, G.N. Cancer microenvironment and inflammation: Role of hyaluronan. Front. Immunol. 2015, 6, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamo, A.; Frusteri, C.; Pallotta, M.T.; Pirali, T.; Sartoris, S.; Ugel, S. Moonlighting Proteins Are Important Players in Cancer Immunology. Front. Immunol. 2020, 11, 613069. [Google Scholar] [CrossRef] [PubMed]
- Nuno-Cabanes, C.; Rodriguez-Navarro, S. The promiscuity of the SAGA complex subunits: Multifunctional or moonlighting proteins? Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194607. [Google Scholar] [CrossRef]
- Prud’homme, B.; Gompel, N.; Carroll, S.B. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 2007, 104 (Suppl. 1), 8605–8612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Chanmee, T.; Itano, N. Hyaluronan: Metabolism and Function. Biomolecules 2020, 10, 1525. [Google Scholar] [CrossRef]
- Triggs-Raine, B.; Natowicz, M.R. Biology of hyaluronan: Insights from genetic disorders of hyaluronan metabolism. World J. Biol. Chem. 2015, 6, 110–120. [Google Scholar] [CrossRef]
- Liu, J.; Yan, W.; Han, P.; Tian, D. The emerging role of KIAA1199 in cancer development and therapy. Biomed. Pharmacother. 2021, 138, 111507. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Yamamoto, H.; Tobisawa, Y.; Irie, F. TMEM2: A missing link in hyaluronan catabolism identified? Matrix Biol. 2019, 78–79, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in wound repair and the “cancerization” of stromal tissues. Biomed. Res. Int. 2014, 2014, 103923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, P.H. Planning, evaluating and vetting receptor signaling studies to assess hyaluronan size-dependence and specificity. Glycobiology 2017, 27, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular hyaluronan: Importance for cellular functions. Semin. Cancer Biol. 2020, 62, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Turley, E.A.; Noble, P.W.; Bourguignon, L.Y. Signaling properties of hyaluronan receptors. J. Biol. Chem. 2002, 277, 4589–4592. [Google Scholar] [CrossRef] [Green Version]
- Avenoso, A.; Bruschetta, G.; D’Ascola, A.; Scuruchi, M.; Mandraffino, G.; Saitta, A.; Campo, S.; Campo, G. Hyaluronan Fragmentation During Inflammatory Pathologies: A Signal that Empowers Tissue Damage. Mini Rev. Med. Chem. 2020, 20, 54–65. [Google Scholar] [CrossRef]
- Jackson, D.G. Hyaluronan in the lymphatics: The key role of the hyaluronan receptor LYVE-1 in leucocyte trafficking. Matrix Biol. 2019, 78–79, 219–235. [Google Scholar] [CrossRef]
- Pratt, R.L. Hyaluronan and the Fascial Frontier. Int. J. Mol. Sci. 2021, 22, 6845. [Google Scholar] [CrossRef]
- Tammi, M.I.; Oikari, S.; Pasonen-Seppanen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated hyaluronan metabolism in the tumor matrix—Causes and consequences. Matrix Biol. 2019, 78–79, 147–164. [Google Scholar] [CrossRef]
- Bollyky, P.L.; Bogdani, M.; Bollyky, J.B.; Hull, R.L.; Wight, T.N. The role of hyaluronan and the extracellular matrix in islet inflammation and immune regulation. Curr. Diab. Rep. 2012, 12, 471–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinelli, F.M.; Vitale, D.L.; Sevic, I.; Alaniz, L. Hyaluronan in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 67–83. [Google Scholar] [PubMed]
- Joy, R.A.; Vikkath, N.; Ariyannur, P.S. Metabolism and mechanisms of action of hyaluronan in human biology. Drug Metab. Pers. Ther. 2018, 33, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Fam, H.; Bryant, J.T.; Kontopoulou, M. Rheological properties of synovial fluids. Biorheology 2007, 44, 59–74. [Google Scholar] [PubMed]
- Gupta, R.C.; Lall, R.; Srivastava, A.; Sinha, A. Hyaluronic Acid: Molecular Mechanisms and Therapeutic Trajectory. Front. Vet. Sci. 2019, 6, 192. [Google Scholar] [CrossRef] [Green Version]
- Rebenda, D.; Vrbka, M.; Cipek, P.; Toropitsyn, E.; Necas, D.; Pravda, M.; Harti, M. On the Dependence of Rheology of Hyaluronic Acid Solutions and Frictional Behavior of Articular Cartilage. Materials 2020, 13, 2659. [Google Scholar] [CrossRef]
- Kavasi, R.M.; Berdiaki, A.; Spyridaki, I.; Corsini, E.; Tsatsakis, A.; Tzanakakis, G.; Nikitovic, D. HA metabolism in skin homeostasis and inflammatory disease. Food Chem. Toxicol. 2017, 101, 128–138. [Google Scholar] [CrossRef]
- Frevert, C.W.; Felgenhauer, J.; Wygrecka, M.; Nastase, M.V.; Schaefer, L. Danger-Associated Molecular Patterns Derived From the Extracellular Matrix Provide Temporal Control of Innate Immunity. J. Histochem. Cytochem. 2018, 66, 213–227. [Google Scholar] [CrossRef]
- Takeo, S.; Fujise, M.; Akiyama, T.; Habuchi, H.; Itano, N.; Matsuo, T.; Aigaki, T.; Kimata, K.; Nakato, H. In vivo hyaluronan synthesis upon expression of the mammalian hyaluronan synthase gene in Drosophila. J. Biol. Chem. 2004, 279, 18920–18925. [Google Scholar] [CrossRef] [Green Version]
- Csoka, A.B.; Stern, R. Hypotheses on the evolution of hyaluronan: A highly ironic acid. Glycobiology 2013, 23, 398–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeAngelis, P.L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002, 268, 317–326. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Mei, L.; Connell, M.; Maxwell, C.A. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather than a Hyaluronan Receptor. Cells 2020, 9, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paschinger, K.; Wilson, I.B.H. Anionic and zwitterionic moieties as widespread glycan modifications in non-vertebrates. Glycoconj J. 2020, 37, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, K.; Maeda, N. Glypicans and Heparan Sulfate in Synaptic Development, Neural Plasticity, and Neurological Disorders. Front. Neural Circuits 2021, 15, 595596. [Google Scholar] [CrossRef]
- Nishihara, S. Glycosyltransferases and transporters that contribute to proteoglycan synthesis in Drosophila: Identification and functional analyses using the heritable and inducible RNAi system. Methods Enzymol. 2010, 480, 323–351. [Google Scholar]
- Stern, R. Go Fly a Chitin: The Mystery of Chitin and Chitinases in Vertebrate Tissues. Front. Biosci. 2017, 22, 580–595. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Itano, N.; Yamada, Y.; Kimata, K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J. Biol. Chem. 2000, 275, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneda, M.; Nakamura, T.; Murai, M.; Wada, H. Evidence for the heparin-binding ability of the ascidian Xlink domain and insight into the evolution of the Xlink domain in chordates. J. Mol. Evol. 2010, 71, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Kawashima, S.; Tanaka, C.; Murai, M.; Yoneda, M.; Putnam, N.H.; Rokhsar, D.S.; Kanehisa, M.; Satoh, N.; Wada, H. Domain shuffling and the evolution of vertebrates. Genome Res. 2009, 19, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Rozbesky, D.; Monistrol, J.; Jain, V.; Hillier, J.; Padilla-Parra, S.; Jones, E.Y. Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility. Structure 2020, 28, 507–515.e5. [Google Scholar] [CrossRef]
- Bernhofer, M.; Dallago, C.; Karl, T.; Satagopam, V.; Heinzinger, M.; Littmann, M.; Olenyi, T.; Qiu, J.; Schutze, K.; Yachdav, G.; et al. PredictProtein-Predicting Protein Structure and Function for 29 Years. Nucleic Acids Res. 2021, 49, W535–W540. [Google Scholar] [CrossRef]
- Yang, B.; Hall, C.L.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a novel heparin binding domain in RHAMM and evidence that it modifies HA mediated locomotion of ras-transformed cells. J. Cell Biochem. 1994, 56, 455–468. [Google Scholar] [CrossRef]
- Tolg, C.; Hamilton, S.R.; Morningstar, L.; Zhang, J.; Zhang, S.; Esguerra, K.V.; Telmer, P.G.; Luyt, L.G.; Harrison, R.; McCarthy, J.B.; et al. RHAMM promotes interphase microtubule instability and mitotic spindle integrity through MEK1/ERK1/2 activity. J. Biol. Chem. 2010, 285, 26461–26474. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yao, L.L.; Li, X.D. Regulation of class V myosin. Cell Mol. Life Sci. 2018, 75, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Abriata, L.A.; Albanesi, D.; Dal Peraro, M.; de Mendoza, D. Signal Sensing and Transduction by Histidine Kinases as Unveiled through Studies on a Temperature Sensor. Acc. Chem. Res. 2017, 50, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.; Vogt, K.; Acosta-Martin, A.E.; Shire, P.; Zeidler, M.; Smythe, E. Integration of JAK/STAT receptor-ligand trafficking, signalling and gene expression in Drosophila melanogaster cells. J. Cell Sci. 2020, 133, jcs246199. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mohan, P.; Jiang, J.; Nemirovsky, O.; He, D.; Fleisch, M.C.; Niederacher, D.; Pilarski, L.M.; Lim, C.J.; Maxwell, C.A. Spatial regulation of Aurora A activity during mitotic spindle assembly requires RHAMM to correctly localize TPX2. Cell Cycle 2014, 13, 2248–2261. [Google Scholar] [CrossRef] [Green Version]
- Meier, C.; Spitschak, A.; Abshagen, K.; Gupta, S.; Mor, J.M.; Wolkenhauer, O.; Haier, J.; Vollmar, B.; Alla, V.; Pützer, B.M. Association of RHAMM with E2F1 promotes tumour cell extravasation by transcriptional up-regulation of fibronectin. J. Pathol. 2014, 234, 351–364. [Google Scholar] [CrossRef]
- Yamasaki, C.; Tashiro, S.; Nishito, Y.; Sueda, T.; Igarashi, K. Dynamic cytoplasmic anchoring of the transcription factor Bach1 by intracellular hyaluronic acid binding protein IHABP. J. Biochem. 2005, 137, 287–296. [Google Scholar] [CrossRef]
- Chen, Y.T.; Chen, Z.; Du, Y.N. Immunohistochemical analysis of RHAMM expression in normal and neoplastic human tissues: A cell cycle protein with distinctive expression in mitotic cells and testicular germ cells. Oncotarget 2018, 9, 20941–20952. [Google Scholar] [CrossRef] [Green Version]
- Savani, R.C. Modulators of inflammation in Bronchopulmonary Dysplasia. Semin. Perinatol. 2018, 42, 459–470. [Google Scholar] [CrossRef]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [Green Version]
- Jain, E.; Bairoch, A.; Duvaud, S.; Phan, I.; Redaschi, N.; Suzek, B.E.; Martin, M.J.; McGarvey, P.; Gasteiger, E. Infrastructure for the life sciences: Design and implementation of the UniProt website. BMC Bioinf. 2009, 10, 136. [Google Scholar] [CrossRef] [Green Version]
- Villegas-Ruiz, V.; Salcedo, M.; Zentella-Dehesa, A.; de Oca, E.V.; Roman-Basaure, E.; Mantilla-Morales, A.; Dávila-Borja, V.M.; Juárez-Méndez, S. A case of cervical cancer expressed three mRNA variant of Hyaluronan-mediated motility receptor. Int. J. Clin. Exp. Pathol. 2014, 7, 2256–2264. [Google Scholar]
- Crainie, M.; Belch, A.R.; Mant, M.J.; Pilarski, L.M. Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: Identification of three distinct RHAMM variants. Blood 1999, 93, 1684–1696. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Rasmussen, E.; Zhan, F.; Keats, J.J.; Adamia, S.; Strachan, E.; Crainie, M.; Walker, R.; Belch, A.R.; Pilarski, L.M.; et al. RHAMM expression and isoform balance predict aggressive disease and poor survival in multiple myeloma. Blood 2004, 104, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Assmann, V.; Jenkinson, D.; Marshall, J.F.; Hart, I.R. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J. Cell Sci. 1999, 112 Pt 22, 3943–3954. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.R.; Fard, S.F.; Paiwand, F.F.; Tolg, C.; Veiseh, M.; Wang, C.; McCarthy, J.B.; Bissell, M.J.; Koropatnick, J.; Turley, E.A. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J. Biol. Chem. 2007, 282, 16667–16680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Line, A.; Slucka, Z.; Stengrevics, A.; Silina, K.; Li, G.; Rees, R.C. Characterisation of tumour-associated antigens in colon cancer. Cancer Immunol. Immunother. 2002, 51, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.C.; Chou, C.K.; Klimstra, D.S.; Varmus, H. Receptor for hyaluronan-mediated motility isoform B promotes liver metastasis in a mouse model of multistep tumorigenesis and a tail vein assay for metastasis. Proc. Natl. Acad. Sci. USA 2011, 108, 16753–16758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Entwistle, J.; Zhang, S.; Yang, B.; Wong, C.; Li, Q.; Hall, C.L.; Mowat, M.; Greenberg, A.H.; Turley, E.A. Characterization of the murine gene encoding the hyaluronan receptor RHAMM. Gene 1995, 163, 233–238. [Google Scholar] [CrossRef]
- Zaman, A.; Cui, Z.; Foley, J.P.; Zhao, H.; Grimm, P.C.; Delisser, H.M.; Savani, R.C. Expression and role of the hyaluronan receptor RHAMM in inflammation after bleomycin injury. Am. J. Respir. Cell Mol. Biol. 2005, 33, 447–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994, 13, 286–296. [Google Scholar] [CrossRef]
- Tolg, C.; Yuan, H.; Flynn, S.M.; Basu, K.; Ma, J.; Tse, K.C.K.; Kowalska, B.; Vullanesku, D.; Cowman, M.K.; McCarthy, J.B.; et al. Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biol. 2017, 63, 117–132. [Google Scholar] [CrossRef]
- Hatano, H.; Shigeishi, H.; Kudo, Y.; Higashikawa, K.; Tobiume, K.; Takata, T.; Kamata, N. RHAMM/ERK interaction induces proliferative activities of cementifying fibroma cells through a mechanism based on the CD44-EGFR. Lab. Investig. 2011, 91, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Chang, M.C.; Zylka, D.; Turley, S.; Harrison, R.; Turley, E.A. The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J. Biol. Chem. 1998, 273, 11342–11348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.M.; Soares da Costa, D.; Paulo, P.M.R.; Reis, R.L.; Pashkuleva, I. Co-localization and crosstalk between CD44 and RHAMM depend on hyaluronan presentation. Acta Biomater. 2021, 119, 114–124. [Google Scholar] [CrossRef]
- Kouvidi, K.; Berdiaki, A.; Tzardi, M.; Karousou, E.; Passi, A.; Nikitovic, D.; Tzanakakis, G.N. Receptor for hyaluronic acid- mediated motility (RHAMM) regulates HT1080 fibrosarcoma cell proliferation via a beta-catenin/c-myc signaling axis. Biochim. Biophys. Acta 2016, 1860, 814–824. [Google Scholar] [CrossRef]
- Blanco, I.; Kuchenbaecker, K.; Cuadras, D.; Wang, X.; Barrowdale, D.; de Garibay, G.R.; Librado, P.; Sánchez-Gracia, A.; Rozas, J.; Bonifaci, N.; et al. Assessing associations between the AURKA-HMMR-TPX2-TUBG1 functional module and breast cancer risk in BRCA1/2 mutation carriers. PLoS ONE 2015, 10, e0120020. [Google Scholar] [CrossRef] [Green Version]
- Tolg, C.; Telmer, P.; Turley, E. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair. PLoS ONE 2014, 9, e88479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziebell, M.R.; Prestwich, G.D. Interactions of peptide mimics of hyaluronic acid with the receptor for hyaluronan mediated motility (RHAMM). J. Comput. Aided Mol. Des. 2004, 18, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Hauser-Kawaguchi, A.; Luyt, L.G.; Turley, E. Design of peptide mimetics to block pro-inflammatory functions of HA fragments. Matrix Biol. 2019, 78–79, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Esguerra, K.V.; Tolg, C.; Akentieva, N.; Price, M.; Cho, C.F.; Lewis, J.D.; McCarthy, J.B.; Turley, E.A.; Luyt, L.G. Identification, design and synthesis of tubulin-derived peptides as novel hyaluronan mimetic ligands for the receptor for hyaluronan-mediated motility (RHAMM/HMMR). Integr. Biol. 2015, 7, 1547–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boittier, E.D.; Gandhi, N.S.; Ferro, V.; Coombe, D.R. Cross-Species Analysis of Glycosaminoglycan Binding Proteins Reveals Some Animal Models Are “More Equal” than Others. Molecules 2019, 24, 924. [Google Scholar] [CrossRef] [Green Version]
- Gulati, K.; Jamsandekar, M.; Poluri, K.M. Mechanistic insights into molecular evolution of species-specific differential glycosaminoglycan binding surfaces in growth-related oncogene chemokines. R. Soc. Open Sci. 2017, 4, 171059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.G.; Triandafillou, C.G.; Huang, T.Y.; Zulueta, M.M.; Banerjee, S.; Dinner, A.R.; Hung, S.H.; Tang, W.J. Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proc. Natl. Acad. Sci. USA 2016, 113, 5000–5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fermas, S.; Gonnet, F.; Sutton, A.; Charnaux, N.; Mulloy, B.; Du, Y.; Baleux, F.; Daniel, R. Sulfated oligosaccharides (heparin and fucoidan) binding and dimerization of stromal cell-derived factor-1 (SDF-1/CXCL 12) are coupled as evidenced by affinity CE-MS analysis. Glycobiology 2008, 18, 1054–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crown, S.E.; Yu, Y.; Sweeney, M.D.; Leary, J.A.; Handel, T.M. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J. Biol. Chem. 2006, 281, 25438–25446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Kim, S.; Liu, V.M.; Sabino, A.; Minkhorst, K.; Yazdani, A.; Turley, E.A. Function-blocking RHAMM peptides attenuate fibrosis and promote anti-fibrotic adipokines in a bleomycin-induced murine model of systemic sclerosis. J. Investig. Dermatol. 2021, 141, 1482–1492.e4. [Google Scholar] [CrossRef] [PubMed]
- Truong, J.L.; Liu, M.; Tolg, C.; Barr, M.; Dai, C.; Raissi, T.C.; Wong, E.; DeLyzer, T.; Yazdani, A.; Turley, E.A. Creating a Favorable Micro-Environment for Fat Grafting in a Novel Model of Radiation Induced Mammary Fat Pad Fibrosis. Plast Reconstr. Surg. 2020, 145, 116–126. [Google Scholar] [CrossRef]
- Cui, Z.; Liao, J.; Cheong, N.; Longoria, C.; Cao, G.; DeLisser, H.M.; Savani, R.C. The Receptor for Hyaluronan-Mediated Motility (CD168) promotes inflammation and fibrosis after acute lung injury. Matrix Biol. 2019, 78–79, 255–271. [Google Scholar] [CrossRef]
- Foley, J.P.; Lam, D.; Jiang, H.; Liao, J.; Cheong, N.; McDevitt, T.M.; Zaman, A.; Wright, J.R.; Savani, R.C. Toll-like receptor 2 (TLR2), transforming growth factor-beta, hyaluronan (HA), and receptor for HA-mediated motility (RHAMM) are required for surfactant protein A-stimulated macrophage chemotaxis. J. Biol. Chem. 2012, 287, 37406–37419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahrami, S.B.; Tolg, C.; Peart, T.; Symonette, C.; Veiseh, M.; Umoh, J.U.; Holdsworth, D.W.; McCarthy, J.B.; Luyt, M.J.; Bissell, M.J.; et al. Receptor for hyaluronan mediated motility (RHAMM/HMMR) is a novel target for promoting subcutaneous adipogenesis. Integr. Biol. 2017, 9, 223–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigeishi, H.; Higashikawa, K.; Takechi, M. Role of receptor for hyaluronan-mediated motility (RHAMM) in human head and neck cancers. J. Cancer Res. Clin. Oncol. 2014, 140, 1629–1640. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Im, J.; Nho, R.S.; Han, Y.H.; Upadhyaya, P.; Kassie, F. Hyaluronan-CD44/RHAMM interaction-dependent cell proliferation and survival in lung cancer cells. Mol. Carcinog. 2019, 58, 321–333. [Google Scholar] [CrossRef]
- Veiseh, M.; Kwon, D.H.; Borowsky, A.D.; Tolg, C.; Leong, H.S.; Lewis, J.D.; Turley, E.A.; Bissell, M.J. Cellular heterogeneity profiling by hyaluronan probes reveals an invasive but slow-growing breast tumor subset. Proc. Natl. Acad. Sci. USA 2014, 111, E1731–E1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, E.A.; Faso, C. The Road Less Traveled? Unconventional Protein Secretion at Parasite-Host Interfaces. Front. Cell Dev. Biol. 2021, 9, 662711. [Google Scholar] [CrossRef]
- Zhang, W.; Lavine, K.J.; Epelman, S.; Evans, S.A.; Weinheimer, C.J.; Barger, P.M.; Mann, D.L. Necrotic myocardial cells release damage-associated molecular patterns that provoke fibroblast activation in vitro and trigger myocardial inflammation and fibrosis in vivo. J. Am. Heart Assoc. 2015, 4, e001993. [Google Scholar] [CrossRef] [Green Version]
- Kahroba, H.; Davatgaran-Taghipour, Y. Exosomal Nrf2: From anti-oxidant and anti-inflammation response to wound healing and tissue regeneration in aged-related diseases. Biochimie 2020, 171–172, 103–109. [Google Scholar] [CrossRef]
- Hascall, V.C.; Majors, A.K.; De La Motte, C.A.; Evanko, S.P.; Wang, A.; Drazba, J.A.; Strong, S.A.; Wight, T.N. Intracellular hyaluronan: A new frontier for inflammation? Biochim. Biophys. Acta 2004, 1673, 3–12. [Google Scholar] [CrossRef]
- Evanko, S.P.; Parks, W.T.; Wight, T.N. Intracellular hyaluronan in arterial smooth muscle cells: Association with microtubules, RHAMM, and the mitotic spindle. J. Histochem. Cytochem. 2004, 52, 1525–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joukov, V.; Groen, A.C.; Prokhorova, T.; Gerson, R.; White, E.; Rodriguez, A.; Walter, J.C.; Livingston, D.M. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 2006, 127, 539–552. [Google Scholar] [CrossRef] [Green Version]
- Tolg, C.; Hamilton, S.R.; Zalinska, E.; McCulloch, L.; Amin, R.; Akentieva, N.; Winnik, F.; Savani, R.; Bagli, D.J.; Luyt, L.G.; et al. A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am. J. Pathol. 2012, 181, 1250–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikonova, A.S.; Astsaturov, I.; Serebriiskii, I.G.; Dunbrack, R.L.; Golemis, E.A. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol. Life Sci. 2013, 70, 661–687. [Google Scholar] [CrossRef] [PubMed]
- Scrofani, J.; Sardon, T.; Meunier, S.; Vernos, I. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol. 2015, 25, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Kroll, T.; Moll, J.; Frappart, L.; Herrlich, P.; Heuer, H.; Ploubidou, A. Spindle Misorientation of Cerebral and Cerebellar Progenitors Is a Mechanistic Cause of Megalencephaly. Stem. Cell Rep. 2017, 9, 1071–1080. [Google Scholar] [CrossRef] [Green Version]
- Silverman-Gavrila, R.V.; Silverman-Gavrila, L.B.; Bilal, K.H.; Bendeck, M.P. Spectrin alpha is important for rear polarization of the microtubule organizing center during migration and spindle pole assembly during division of neointimal smooth muscle cells. Cytoskeleton 2015, 72, 157–170. [Google Scholar] [CrossRef]
- Dunsch, A.K.; Hammond, D.; Lloyd, J.; Schermelleh, L.; Gruneberg, U.; Barr, F.A. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J. Cell Biol. 2012, 198, 1039–1054. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Moll, J.; Winkler, A.; Frappart, L.; Brunet, S.; Hamann, J.; Kroll, T.; Verlhac, M.H.; Heuer, H.; Herrlich, P.; et al. RHAMM deficiency disrupts folliculogenesis resulting in female hypofertility. Biol. Open 2015, 4, 562–571. [Google Scholar] [CrossRef] [Green Version]
- Eibes, S.; Gallisa-Sune, N.; Rosas-Salvans, M.; Martinez-Delgado, P.; Vernos, I.; Roig, J. Nek9 Phosphorylation Defines a New Role for TPX2 in Eg5-Dependent Centrosome Separation before Nuclear Envelope Breakdown. Curr. Biol. 2018, 28, 121–129.e4. [Google Scholar] [CrossRef] [Green Version]
- Tolg, C.; Poon, R.; Fodde, R.; Turley, E.A.; Alman, B.A. Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor). Oncogene 2003, 22, 6873–6882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bost, F.; Aouadi, M.; Caron, L.; Even, P.; Belmonte, N.; Prot, M.; Dani, C.; Hofman, P.; Pagès, G.; Pouysségur, J.; et al. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes 2005, 54, 402–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Bello, B.; Rangel-Garcia, C.I.; Salvador, C.; Carrisoza-Gaytan, R.; Escobar, L.I. The polarization of the G-protein activated potassium channel GIRK5 to the vegetal pole of Xenopus laevis oocytes is driven by a di-leucine motif. PLoS ONE 2013, 8, e64096. [Google Scholar] [CrossRef]
- Murrell-Lagnado, R.D.; Frick, M. P2X4 and lysosome fusion. Curr. Opin. Pharmacol. 2019, 47, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jin, S.; Cao, Y.; Xu, J.; Zhu, S.; Li, Z. Emodin regulates cell cycle of non-small lung cancer (NSCLC) cells through hyaluronan synthase 2 (HA2)-HA-CD44/receptor for hyaluronic acid-mediated motility (RHAMM) interaction-dependent signaling pathway. Cancer Cell Int. 2021, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, C.; Boopathi, E.; Liu, Y.; Haber, A.; Ertel, A.; Bhardwaj, A.; Addya, S.; Williams, N.; Ciment, S.J.; Cotzia, P.; et al. RB Loss Promotes Prostate Cancer Metastasis. Cancer Res. 2017, 77, 982–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, J.I.; Camenisch, T.D.; Stevens, M.V.; Sands, B.J.; McDonald, J.; Schroeder, J.A. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005, 65, 6755–6763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Thor, A.D.; Moore, D.H.; Zhao, Y.; Kerschmann, R.; Stern, R.; Watson, P.H.; Turley, E.A. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin. Cancer Res. 1998, 4, 567–576. [Google Scholar] [PubMed]
- Schutze, A.; Vogeley, C.; Gorges, T.; Twarock, S.; Butschan, J.; Babayan, A.; Klein, D.; Knauer, S.K.; Metzen, E.; Müller, V.; et al. RHAMM splice variants confer radiosensitivity in human breast cancer cell lines. Oncotarget 2016, 7, 21428–21440. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Liu, X.; Lv, M.; Sun, E.; Lu, X.; Lu, C. Genes That Predict Poor Prognosis in Breast Cancer via Bioinformatical Analysis. Biomed. Res. Int. 2021, 2021, 6649660. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.Z. Adhesion molecules--The lifelines of multiple myeloma cells. Semin. Cancer Biol. 2010, 20, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.W.; Wang, S.; Wu, Z.Z.; Yang, Q.C.; Chen, D.R.; Wan, S.C.; Sun, Z.J. Overexpression of CD168 is related to poor prognosis in oral squamous cell carcinoma. Oral Dis. 2021. [Google Scholar] [CrossRef]
- Buttermore, S.T.; Hoffman, M.S.; Kumar, A.; Champeaux, A.; Nicosia, S.V.; Kruk, P.A. Increased RHAMM expression relates to ovarian cancer progression. J. Ovarian Res. 2017, 10, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Narula, N.; Azzopardi, S.; Smith, R.S.; Nasar, A.; Altorki, N.K.; Mittal, V.; Somwar, R.; Stiles, B.M.; Du, Y.C.N. Expression of the receptor for hyaluronic acid mediated motility (RHAMM) is associated with poor prognosis and metastasis in non-small cell lung carcinoma. Oncotarget 2016, 7, 39957–39969. [Google Scholar] [CrossRef] [Green Version]
- Rizzardi, A.E.; Vogel, R.I.; Koopmeiners, J.S.; Forster, C.L.; Marston, L.O.; Rosener, N.K.; Akentieva, N.; Price, M.A.; Metzger, G.J.; Warlick, C.A.; et al. Elevated hyaluronan and hyaluronan-mediated motility receptor are associated with biochemical failure in patients with intermediate-grade prostate tumors. Cancer 2014, 120, 1800–1809. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Gao, C.; Liu, Y.; Wen, Y.; Hong, X.; Huang, Z. Bioinformatics Analysis of Prognostic miRNA Signature and Potential Critical Genes in Colon Cancer. Front. Genet. 2020, 11, 478. [Google Scholar] [CrossRef]
- Lu, X.Q.; Zhang, J.Q.; Zhang, S.X.; Qiao, J.; Qiu, M.T.; Liu, X.R.; Chen, X.X.; Gao, C.; Zhang, H.H. Identification of novel hub genes associated with gastric cancer using integrated bioinformatics analysis. BMC Cancer 2021, 21, 697. [Google Scholar] [CrossRef]
- Levy, P.; Vidaud, D.; Leroy, K.; Laurendeau, I.; Wechsler, J.; Bolasco, G.; Parfait, B.; Wolkenstein, P.; Vidaud, M.; Bièche, I. Molecular profiling of malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1, based on large-scale real-time RT-PCR. Mol. Cancer 2004, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, Inflammation, and Breast Cancer Progression. Front. Immunol. 2015, 6, 236. [Google Scholar] [CrossRef]
- Gurevich, E.V.; Gurevich, V.V. Beyond traditional pharmacology: New tools and approaches. Br. J. Pharmacol. 2015, 172, 3229–3241. [Google Scholar] [CrossRef] [Green Version]
- Savani, R.C.; Hou, G.; Liu, P.; Wang, C.; Simons, E.; Grimm, P.C.; Stern, R.; Greenberg, A.H.; DeLisser, H.M.; Khalil, N. A role for hyaluronan in macrophage accumulation and collagen deposition after bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 2000, 23, 475–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.M.; Chen, Y.; Chen, J.; Yang, S.; Gao, F.; Underhill, C.B.; Creswell, K.; Zhang, L. A peptide with three hyaluronan binding motifs inhibits tumor growth and induces apoptosis. Cancer Res. 2003, 63, 5685–5690. [Google Scholar] [PubMed]
- Akentieva, N.P.; Shushanov, S.S. Inhibitory effect of RHAMM-target peptides on invasion of breast cancer cells. Vopr. Onkol. 2016, 62, 831–838. [Google Scholar] [PubMed]
- Akentieva, N.P.; Shushanov, S.S. RHAMM (receptor hyaluronan-mediated motility)-target peptides induce apoptosis in prostate cancer cells. Vopr. Onkol. 2016, 62, 514–518. [Google Scholar] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messam, B.J.; Tolg, C.; McCarthy, J.B.; Nelson, A.C.; Turley, E.A. RHAMM Is a Multifunctional Protein That Regulates Cancer Progression. Int. J. Mol. Sci. 2021, 22, 10313. https://doi.org/10.3390/ijms221910313
Messam BJ, Tolg C, McCarthy JB, Nelson AC, Turley EA. RHAMM Is a Multifunctional Protein That Regulates Cancer Progression. International Journal of Molecular Sciences. 2021; 22(19):10313. https://doi.org/10.3390/ijms221910313
Chicago/Turabian StyleMessam, Britney J., Cornelia Tolg, James B. McCarthy, Andrew C. Nelson, and Eva A. Turley. 2021. "RHAMM Is a Multifunctional Protein That Regulates Cancer Progression" International Journal of Molecular Sciences 22, no. 19: 10313. https://doi.org/10.3390/ijms221910313






