A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides’ Precursors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of LTL1 from L. theobromae with Zymography-LC-MS
2.2. Expression of Lipolytic Enzyme in Fed-Batch Fermentation
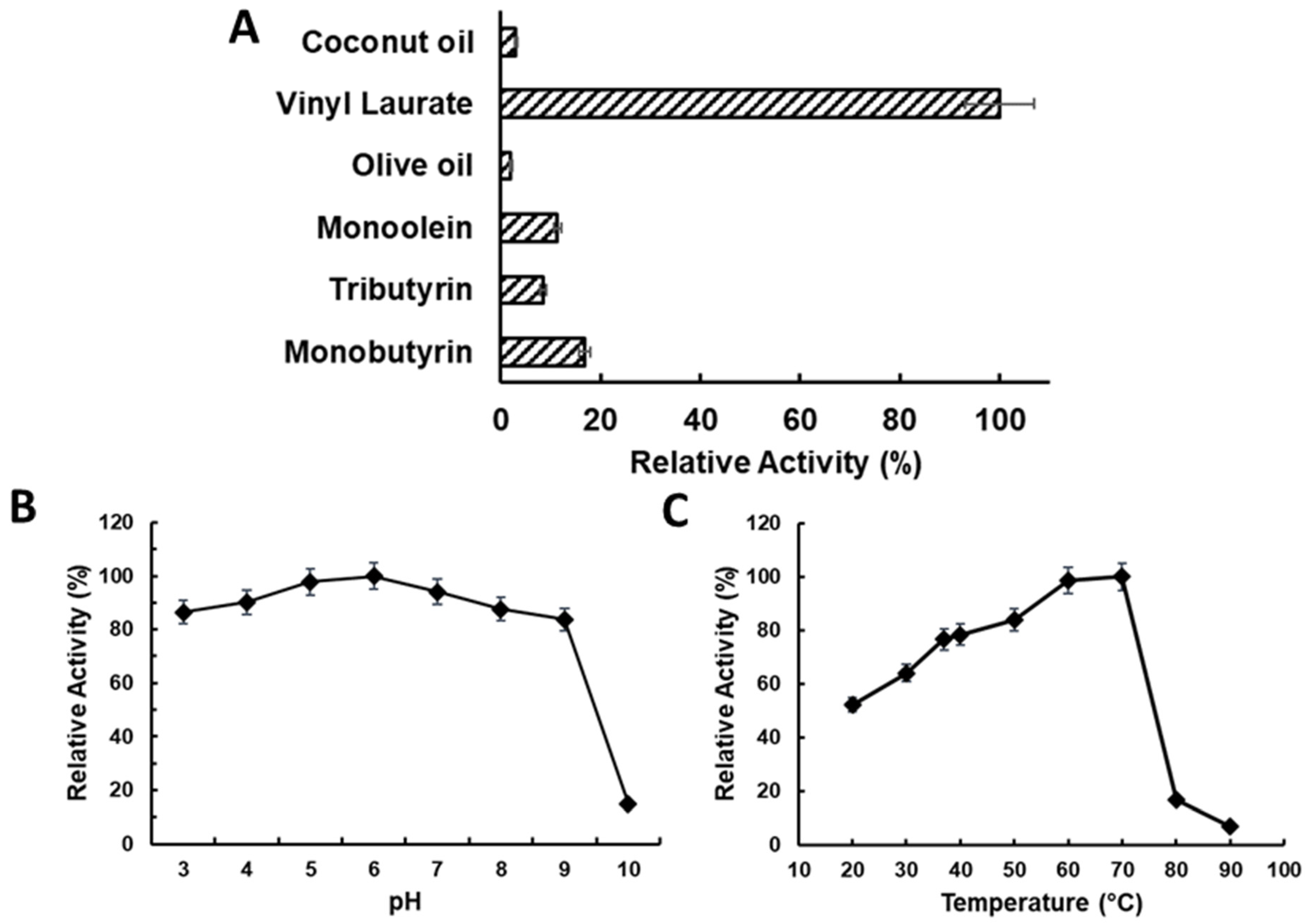
2.3. Characterising LTL1 Substrate, pH, and Temperature Specificities
2.4. LTL1 Efficiently Hydrolyses C8-C10 ME
2.5. Protein X-ray Crystallography of LTL1
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Detection and Identification of LTL1 with Zymography-LC-MS
3.3. Strain Construction and Transformation into Expression Strain
3.4. Fed-batch Fermentation Conditions
3.5. Enzyme Activity Characterisation Conditions
3.6. Enzyme Preparation for Protein X-ray Crystallisation
3.7. Protein X-ray Crystallisation Conditions
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avgerinos, K.I.; Egan, J.M.; Mattson, M.P.; Kapogiannis, D. Medium Chain Triglycerides induce mild ketosis and may improve cognition in Alzheimer’s disease. A systematic review and meta-analysis of human studies. Ageing Res. Rev. 2020, 58, 101001. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, Y.; Zhang, X.; Liu, L.; Zhou, B.; Mo, R.; Li, Y.; Li, H.; Li, F.; Tao, Y.; et al. Medium-chain triglycerides improved cognition and lipid metabolomics in mild to moderate Alzheimer’s disease patients with APOE4: A double-blind, randomized, placebo-controlled crossover trial. Clin. Nutr. 2020, 39, 2092–2105. [Google Scholar] [CrossRef]
- Ota, M.; Matsuo, J.; Ishida, I.; Takano, H.; Yokoi, Y.; Hori, H.; Yoshida, S.; Ashida, K.; Nakamura, K.; Takahashi, T.; et al. Effects of a medium-chain triglyceride-based ketogenic formula on cognitive function in patients with mild-to-moderate Alzheimer’s disease. Neurosci. Lett. 2019, 690, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bemis, M.; Desilets, A.R. Role of Medium Chain Triglycerides (Axona®) in the Treatment of Mild to Moderate Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Dement. 2014, 29, 409–414. [Google Scholar] [CrossRef]
- Croteau, E.; Castellano, C.-A.; Richard, M.A.; Fortier, M.; Nugent, S.; Lepage, M.; Duchesne, S.; Whittingstall, K.; Turcotte, É.E.; Bocti, C.; et al. Ketogenic Medium Chain Triglycerides Increase Brain Energy Metabolism in Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 64, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.C.; Babayan, V.K. Medium-chain triglycerides: An update. Am. J. Clin. Nutr. 1982, 36, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Marten, B.; Pfeuffer, M.; Schrezenmeir, J. Medium-chain triglycerides. Int. Dairy J. 2006, 16, 1374–1382. [Google Scholar] [CrossRef]
- Xue, C.; Liu, Y.; Wang, J.; Zhang, R.; Zhang, Y.; Zhang, J.; Zhang, Y.; Zheng, Z.; Yu, X.; Jing, H.; et al. Consumption of medium- and long-chain triacylglycerols decreases body fat and blood triglyceride in Chinese hypertriglyceridemic subjects. Eur. J. Clin. Nutr. 2009, 63, 879–886. [Google Scholar] [CrossRef] [Green Version]
- Mett, J.; Müller, U. The medium-chain fatty acid decanoic acid reduces oxidative stress levels in neuroblastoma cells. Sci. Rep. 2021, 11, 6135. [Google Scholar] [CrossRef]
- Langone, M.A.P.; Sant’Anna, G.L. Process development for production of medium chain triglycerides using immobilized lipase in a solvent-free system. Appl. Biochem. Biotechnol. 2002, 98, 997–1008. [Google Scholar] [CrossRef]
- Ooi, T.L.; Yong, K.C.; Hazimah, A.H.; Dzulkefly, K.; Yunus, W.M.Z.W. Glycerol Residue—A Rich Source of Glycerol and Medium Chain Fatty Acids. J. Oleo Sci. 2004, 53, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Shirazi, Y.; Viamajala, S.; Varanasi, S. High-yield production of fuel- and oleochemical-precursors from triacylglycerols in a novel continuous-flow pyrolysis reactor. Appl. Energy 2016, 179, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Casas-Godoy, L.; Duquesne, S.; Bordes, F.; Sandoval, G.; Marty, A. Lipases: An Overview. In Lipases and Phospholipases: Methods and Protocols; Sandoval, G., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 3–30. [Google Scholar]
- Kundys, A.; Białecka-Florjańczyk, E.; Fabiszewska, A.; Małajowicz, J. Candida antarctica Lipase B as Catalyst for Cyclic Esters Synthesis, Their Polymerization and Degradation of Aliphatic Polyesters. J. Polym. Environ. 2018, 26, 396–407. [Google Scholar] [CrossRef]
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique Biocatalysts from a Unique Origin. Org. Process Res. Dev. 2002, 6, 446–451. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal. B Enzym. 2010, 64, 1–22. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ayub, M.A.Z. Effects of the combined use of Thermomyces lanuginosus and Rhizomucor miehei lipases for the transesterification and hydrolysis of soybean oil. Process. Biochem. 2011, 46, 682–688. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Ng, A.M.J.; Zhang, H.; Nguyen, G.K.T. Zymography for Picogram Detection of Lipase and Esterase Activities. Molecules 2021, 26, 1542. [Google Scholar] [CrossRef]
- Yan, J.Y.; Zhao, W.S.; Chen, Z.; Xing, Q.K.; Zhang, W.; Chethana, K.W.T.; Xue, M.F.; Xu, J.P.; Phillips, A.J.L.; Wang, Y.; et al. Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res. 2018, 25, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Invitrogen Pichia Fermentation Process Guidelines. Available online: https://tools.thermofisher.com/content/sfs/manuals/pichiaferm_prot.pdf (accessed on 28 June 2021).
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Bisong, E. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Apress: Berkeley, CA, USA, 2019; pp. 59–64. [Google Scholar]
- Holm, L.; Rosenström, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Using Dali for Protein Structure Comparison. Methods Mol. Biol. 2020, 2112, 29–42. [Google Scholar] [PubMed]
- Huang, W.; Lan, D.; Popowicz, G.M.; Zak, K.M.; Zhao, Z.; Yuan, H.; Yang, B.; Wang, Y. Structure and characterization of Aspergillus fumigatus lipase B with a unique, oversized regulatory subdomain. FEBS J. 2019, 286, 2366–2380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation1. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, R.; Pan, S.; Peng, Z.; Zhang, Y.; Yang, J. mTM-align: A server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Res. 2018, 46, W380–W386. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. SDS-Polyacrylamide Gel Electrophoresis of Proteins. CSH Protoc. 2006, 2006. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Michalski, A.; Damoc, E.; Hauschild, J.P.; Lange, O.; Wieghaus, A.; Makarov, A.; Nagaraj, N.; Cox, J.; Mann, M.; Horning, S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteom. 2011, 10, M111.011015. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Letchworth, G.J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. BioTechniques 2004, 36, 152–154. [Google Scholar] [CrossRef]
- AOCS Official Method Cd 3d-63: Acid Value of Fats and Oils, 7th ed.; American Oil Chemist’s Society: Urbana, IL, USA, 2020.
- Cowieson, N.P.; Aragao, D.; Clift, M.; Ericsson, D.J.; Gee, C.; Harrop, S.J.; Mudie, N.; Panjikar, S.; Price, J.R.; Riboldi-Tunnicliffe, A.; et al. MX1: A bending-magnet crystallography beamline serving both chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2015, 22, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.L.; Zhang, H.F.; Ye, W.; Ng, M.J.A.; Nguyen, K.T.G. Polypeptides with Lipase Activity and Uses Thereof. U.S. Patent Publication No. 20210198704, 1 July 2021. [Google Scholar]



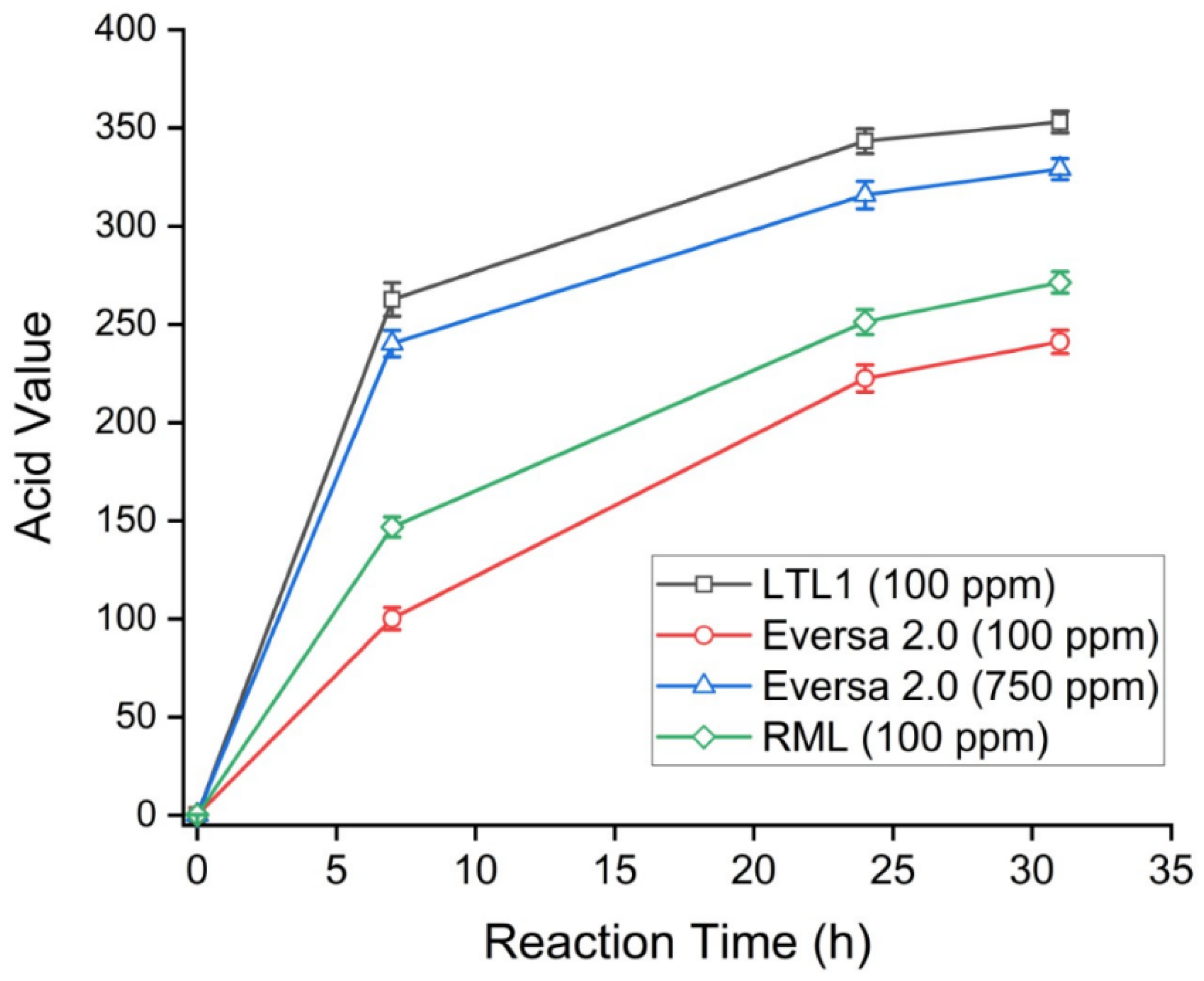

| Time (h) | Acid Value (Max Value = 365) | |||
|---|---|---|---|---|
| LTL1 (100 ppm) | Eversa 2.0 (100 ppm) | Eversa 2.0 (750 ppm) | RML (100 ppm) | |
| 0 | 0.26 (±0.04) | 0.26 (±0.04) | 0.26 (±0.04) | 0.26 (±0.04) |
| 7 | 262.62 (±8.56) | 100.15 (±5.66) | 240.25 (±6.73) | 146.84 (±5.21) |
| 24 | 343.26 (±6.32) | 222.52 (±6.89) | 315.83 (±7.02) | 251.21 (±6.39) |
| 31 | 353.02 (±5.53) | 241.16 (±5.95) | 329.01 (±5.36) | 271.34 (±5.47) W |
| PDB ID | 7V6D |
|---|---|
| Data collection | |
| Wavelength | 0.95373 |
| Resolution range | 29.49-2.50 (2.59-2.50) |
| Space group | I 2 2 2 |
| Unit cell | 65.43 162.64 166.67 90 90 90 |
| Total reflections | 426,375 (42,857) |
| Unique reflections | 31,232 (3060) |
| Multiplicity | 13.7 (14.0) |
| Completeness (%) | 99.88 (100.00) |
| Mean I/sigma(I) | 12.04 (2.20) |
| Wilson B-factor | 36.46 |
| R-merge | 0.189 (1.180) |
| R-meas | 0.196 (1.220) |
| R-pim | 0.053 (0.325) |
| CC1/2 | 0.997 (0.844) |
| CC * | 0.999 (0.957) |
| Refinement | |
| Reflections used in refinement | 31,223 (3060) |
| Reflections used for R-free | 1555 (159) |
| R-work | 0.195 (0.268) |
| R-free | 0.242 (0.322) |
| CCwork | 0.951 (0.914) |
| CCfree | 0.929 (0.794) |
| Number of non-hydrogen atoms | 5570 |
| macromolecules | 5422 |
| ligands | 30 |
| solvent | 118 |
| Protein residues | 733 |
| RMS (bonds) | 0.007 |
| RMS (angles) | 0.99 |
| Validation | |
| Ramachandran favoured (%) | 97.26 |
| Ramachandran allowed (%) | 2.74 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 1.37 |
| Clash score | 3.82 |
| Average B-factor | 43.73 |
| macromolecules | 43.76 |
| Ligands | 48.06 |
| Solvent | 41.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, A.M.J.; Yang, R.; Zhang, H.; Xue, B.; Yew, W.S.; Nguyen, G.K.T. A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides’ Precursors. Int. J. Mol. Sci. 2021, 22, 10339. https://doi.org/10.3390/ijms221910339
Ng AMJ, Yang R, Zhang H, Xue B, Yew WS, Nguyen GKT. A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides’ Precursors. International Journal of Molecular Sciences. 2021; 22(19):10339. https://doi.org/10.3390/ijms221910339
Chicago/Turabian StyleNg, Andre Mong Jie, Renliang Yang, Hongfang Zhang, Bo Xue, Wen Shan Yew, and Giang Kien Truc Nguyen. 2021. "A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides’ Precursors" International Journal of Molecular Sciences 22, no. 19: 10339. https://doi.org/10.3390/ijms221910339







