Prognostic and Predictive Biomarkers in Gliomas
Abstract
1. Introduction
2. The 2021 WHO Classification of Tumors of the Central Nervous System
3. IDH1 and IDH2
Positive Prognostic Factors
4. H3F3A
Negative Prognostic Factor
5. ATRX
Prognostic Factor
6. TERT
Negative Prognostic Factor
7. CDKN2A
Negative Prognostic Factor
8. 1p/19q Codeletion
8.1. Classifying Marker
8.2. Prognostic Factor
8.3. Predictive Factor
9. Chromosome 7 Gain and Chromosome 10 Loss
9.1. Negative Prognostic Factor
9.2. Positive Predictive Marker
10. MYB
Positive Prognostic Factor
11. MN1
Positive Prognostic Factor
12. MAPK Pathway
Positive Prognostic Factor
13. EGFR
13.1. Negative Prognostic Factor
13.2. Promising Predictive Factor
14. MGMT
14.1. Positive Predictive Factor
14.2. Positive Prognostic Factor
15. Mismatch Repair
15.1. Hypermutation Phenotype
15.2. Questionable Predictive Value for Immune Checkpoint Inhibitors (ICI)
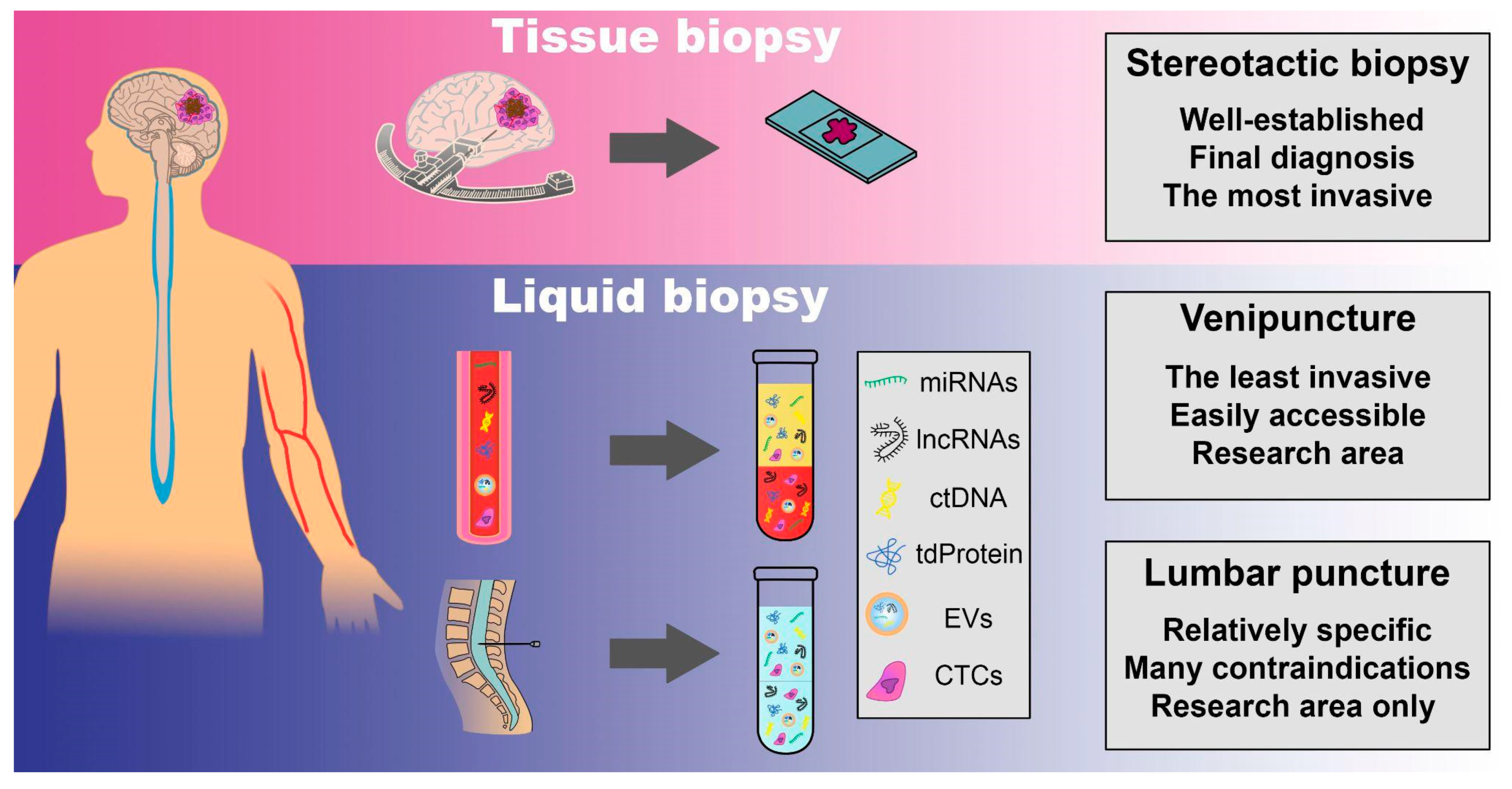
16. Liquid Biopsies
17. Circulating Tumor DNA
18. MicroRNAs
18.1. Potential Classification Marker
18.2. Potential Prognostic and Predictive Markers
19. Long Non-Coding RNA
19.1. Prognostic Markers
19.2. Predictive Markers
20. Tumor-Derived Proteins
20.1. Prognostic Markers
20.2. Predictive Markers
21. Extracellular Vesicles
22. Circulating Tumor Cells
23. Conclusions
23.1. Prognostic Markers
23.2. Predictive Markers
23.3. Liquid Biopsies
Author Contributions
Funding
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Schwartzbaum, J.A.; Fisher, J.L.; Aldape, K.D.; Wrensch, M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006, 2, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef]
- McNeill, K.A. Epidemiology of Brain Tumors. Neurol. Clin. 2016, 34, 981–998. [Google Scholar] [CrossRef]
- Hansen, S.; Rasmussen, B.K.; Laursen, R.J.; Kosteljanetz, M.; Schultz, H.; Nørgård, B.M.; Guldberg, R.; Gradel, K.O. Treatment and survival of glioblastoma patients in Denmark: The Danish Neuro-Oncology Registry 2009–2014. J. Neuro-Oncol. 2018, 139, 479–489. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Sant, M.; Minicozzi, P.; Lagorio, S.; Johannesen, T.B.; Marcos-Gragera, R.; Francisci, S. The EUROCARE Working Group Survival of European patients with central nervous system tumors. Int. J. Cancer 2011, 131, 173–185. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- De Vleeschouwer, S.; Bergers, G. Glioblastoma: To Target the Tumor Cell or the Microenvironment? In Glioblastoma; de Vleeschouwer, S., Ed.; Codon Publications: Brisbane, QLD, Australia, 2017; ISBN 9780994438126. [Google Scholar]
- Zhu, P.; Zhu, J.-J. Tumor treating fields: A novel and effective therapy for glioblastoma: Mechanism, efficacy, safety and future perspectives. Chin. Clin. Oncol. 2017, 6, 41. [Google Scholar] [CrossRef]
- Gera, N.; Yang, A.; Holtzman, T.S.; Lee, S.X.; Wong, E.T.; Swanson, K.D. Tumor Treating Fields Perturb the Localization of Septins and Cause Aberrant Mitotic Exit. PLoS ONE 2015, 10, e0125269. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.; Patel, C.B.; Pohling, C.; Young, C.; Song, J.; Flores, T.A.; Zeng, Y.; Joubert, L.-M.; Arami, H.; Natarajan, A.; et al. Tumor treating fields increases membrane permeability in glioblastoma cells. Cell Death Discov. 2018, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs. Maintenance Temozolomide Alone on Survival in Patients with Glioblastoma. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Brandsma, D.; Stalpers, L.; Taal, W.; Sminia, P.; Bent, M.J.V.D. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008, 9, 453–461. [Google Scholar] [CrossRef]
- De Wit, M.C.; De Bruin, H.G.; Eijkenboom, W.; Smitt, P.A.S.; Bent, M.V.D. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology 2004, 63, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Aldape, K.; Brat, D.J.; Capper, D.; Cree, I.A.; Eberhart, C.; Figarella-Branger, D.; Fouladi, M.; Fuller, G.N.; et al. cIMPACT-NOW update 6: New entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020, 30, 844–856. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 Mutations in Gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 1–7. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Tang, K.; Liang, T.-Y.; Zhang, W.-Z.; Li, J.-Y.; Wang, W.; Hu, H.-M.; Li, M.-Y.; Wang, H.-Q.; He, X.-Z.; et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J. Exp. Clin. Cancer Res. 2016, 35, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ye, D.; Guan, K.-L.; Xiong, Y. IDH1 and IDH2 Mutations in Tumorigenesis: Mechanistic Insights and Clinical Perspectives. Clin. Cancer Res. 2012, 18, 5562–5571. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Ježek, P. 2-Hydroxyglutarate in Cancer Cells. Antioxid. Redox Signal. 2020, 33, 903–926. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1. Science 2009, 324, 261–265. [Google Scholar] [CrossRef]
- Lu, C.; Ward, P.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Atai, N.A.; Lamba, S.E.; Jonker, A.; Rijkeboer, D.; Bosch, K.S.; Tigchelaar, W.; Troost, D.; Vandertop, W.P.; Bardelli, A.; et al. The prognostic IDH1 R132 mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010, 119, 487–494. [Google Scholar] [CrossRef]
- Metellus, P.; Coulibaly, B.; Colin, C.; De Paula, A.M.; Vasiljevic, A.; Taieb, D.; Barlier, A.; Boisselier, B.; Mokhtari, K.; Wang, X.W.; et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010, 120, 719–729. [Google Scholar] [CrossRef]
- Brat, D.J.; Aldape, K.; Colman, H.; Holland, E.C.; Louis, D.N.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.K.; Perry, A.; Reifenberger, G.; Stupp, R.; et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018, 136, 805–810. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.W.; Aldape, K.D.; Yung, W.K.A.; Salama, S.R.; Cooper, L.A.D.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Ichimura, K.; Pearson, D.M.; Kocialkowski, S.; Baäcklund, L.M.; Chan, R.; Jones, D.T.W.; Collins, V.P. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-Oncology 2009, 11, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Songtao, Q.; Lei, Y.; Si, G.; Yanqing, D.; Huixia, H.; Xuelin, Z.; Lanxiao, W.; Fei, Y. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2011, 103, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Sabha, N.; Knobbe, C.B.; Maganti, M.; Al Omar, S.; Bernstein, M.; Cairns, R.; Cako, B.; von Deimling, A.; Capper, D.; Mak, T.W.; et al. Analysis of IDH mutation, 1p/19q deletion, and PTEN loss delineates prognosis in clinical low-grade diffuse gliomas. Neuro-Oncology 2014, 16, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Yin, L.; Li, S.; Han, S.; Song, G.; Liu, N.; Yan, C. Prognostic significance of IDH mutation in adult low-grade gliomas: A meta-analysis. J. Neuro-Oncology 2013, 113, 277–284. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, W.; Wang, Y.; Peng, X.; Chen, B.; Qiu, X.; Li, G.; Li, S.; Wu, C.; Yao, K.; et al. IDH mutation and MGMT promoter methylation in glioblastoma: Results of a prospective registry. Oncotarget 2015, 6, 40896–40906. [Google Scholar] [CrossRef]
- Lewandowska, M.A.; Furtak, J.; Szylberg, T.; Roszkowski, K.; Windorbska, W.; Rytlewska, J.; Jozwicki, W. An analysis of the prognostic value of IDH1 (isocitrate dehydrogenase 1) mutation in Polish glioma patients. Mol. Diagn. Ther. 2013, 18, 45–53. [Google Scholar] [CrossRef]
- Zou, P.; Xu, H.; Chen, P.; Yan, Q.; Zhao, L.; Zhao, P.; Gu, A. IDH1/IDH2 Mutations Define the Prognosis and Molecular Profiles of Patients with Gliomas: A Meta-Analysis. PLoS ONE 2013, 8, e68782. [Google Scholar] [CrossRef]
- Dai, Y.; Ning, X.; Han, G.; Li, W. Assessment of the Association between Isocitrate Dehydrogenase 1 Mutation and Mortality Risk of Glioblastoma Patients. Mol. Neurobiol. 2015, 53, 1501–1508. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Verbaan, D.; Lamba, S.E.; Zanon, C.; Jeuken, J.W.; Boots-Sprenger, S.H.; Wesseling, P.; Hulsebos, T.J.; Troost, D.; Van Tilborg, A.A.; et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro-Oncology 2014, 16, 1263–1273. [Google Scholar] [CrossRef]
- Cubuk, C.; Hidalgo, M.R.; Amadoz, A.; Pujana, M.A.; Mateo, F.; Herranz, C.; Carbonell-Caballero, J.; Dopazo, J. Gene Ex-pression Integration into Pathway Modules Reveals a Pan-Cancer Metabolic Landscape. Cancer Res. 2018, 78, 6059–6072. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants—Ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010, 11, 264–275. [Google Scholar] [CrossRef]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [CrossRef]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.-A.; Jones, D.T.W.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Khuong-Quang, D.-A.; Buczkowicz, P.; Rakopoulos, P.; Liu, X.-Y.; Fontebasso, A.M.; Bouffet, E.; Bartels, U.; Albrecht, S.; Schwartzentruber, J.; Letourneau, L.; et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012, 124, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.-Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.-A.K.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Hamisch, C.; Kickingereder, P.; Fischer, M.; Simon, T.; Ruge, M.I. Update on the diagnostic value and safety of stereotactic biopsy for pediatric brainstem tumors: A systematic review and meta-analysis of 735 cases. J. Neurosurg. Pediatr. 2017, 20, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Ryzhova, M.; Hovestadt, V.; Bender, S.; Sturm, D.; Capper, D.; Meyer, J.; Schrimpf, D.; Kool, M.; Northcott, P.A.; et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015, 129, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Gerges, N.; Korshunov, A.; Sabha, N.; Khuong-Quang, D.-A.; Fontebasso, A.M.; Fleming, A.; Hadjadj, D.; Schwartzentruber, J.; Majewski, J.; et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012, 124, 615–625. [Google Scholar] [CrossRef]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; De Lange, T.; De, S.; Petrini, J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef]
- Kannan, K.; Inagaki, A.; Silber, J.; Gorovets, D.; Zhang, J.; Kastenhuber, E.R.; Heguy, A.; Petrini, J.H.; Chan, T.A.; Huse, J.T. Whole exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget 2012, 3, 1194–1203. [Google Scholar] [CrossRef]
- Jiao, Y.; Killela, P.J.; Reitman, Z.; Rasheed, B.A.; Heaphy, C.M.; De Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Shinjo, S.; Marie, S.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Pekmezci, M.; Rice, T.; Molinaro, A.M.; Walsh, K.; Decker, P.A.; Hansen, H.; Sicotte, H.; Kollmeyer, T.M.; McCoy, L.S.; Sarkar, G.; et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: Additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017, 133, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Figarella-Branger, D.; Mokhtari, K.; Dehais, C.; Jouvet, A.; Uro-Coste, E.; Colin, C.; Carpentier, C.; Forest, F.; Maurage, C.-A.; Vignaud, J.-M.; et al. Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations. Neuro-Oncology 2014, 16, 1244–1254. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, W.; Zhao, H. Loss of heterozygosity 1p/19q and survival in glioma: A meta-analysis. Neuro-Oncology 2013, 16, 103–112. [Google Scholar] [CrossRef]
- Ryall, S.; Krishnatry, R.; Arnoldo, A.; Buczkowicz, P.; Mistry, M.; Siddaway, R.; Ling, C.; Pajovic, S.; Yu, M.; Rubin, J.B.; et al. Targeted detection of genetic alterations reveal the prognostic impact of H3K27M and MAPK pathway aberrations in paediatric thalamic glioma. Acta Neuropathol. Commun. 2016, 4, 1–10. [Google Scholar] [CrossRef]
- Meng, W.; Jiang, Y.; Ma, J. Is the prognostic significance of O6-methylguanine-DNA methyltransferase promoter methylation equally important in glioblastomas of patients from different continents? A systematic review with meta-analysis. Cancer Manag. Res. 2017, 9, 411–425. [Google Scholar] [CrossRef]
- Zhao, Y.-H.; Wang, X.; Cao, C.-J.; Weng, H.; Xu, C.-S.; Li, K.; Li, J.-L.; Lan, J.; Zeng, X.-T.; Li, Z.-Q. The Clinical Significance of O6-Methylguanine-DNA Methyltransferase Promoter Methylation Status in Adult Patients with Glioblastoma: A Meta-analysis. Front. Neurol. 2018, 9, 127. [Google Scholar] [CrossRef]
- Vuong, H.G.; Nguyen, T.Q.; Ngo, T.N.M.; Nguyen, H.C.; Fung, K.-M.; Dunn, I.F. The interaction between TERT promoter mutation and MGMT promoter methylation on overall survival of glioma patients: A meta-analysis. BMC Cancer 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Arita, H.; Yamasaki, K.; Matsushita, Y.; Nakamura, T.; Shimokawa, A.; Takami, H.; Tanaka, S.; Mukasa, A.; Shirahata, M.; Shimizu, S.; et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 2016, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Altibi, A.M.; Duong, U.N.; Ngo, H.T.; Pham, T.Q.; Chan, A.K.-Y.; Park, C.-K.; Fung, K.-M.; Hassell, L. TERT promoter mutation and its interaction with IDH mutations in glioma: Combined TERT promoter and IDH mutations stratifies lower-grade glioma into distinct survival subgroups—A meta-analysis of aggregate data. Crit. Rev. Oncol. 2017, 120, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.J.; Nabavizadeh, S.A.; Till, J.; Abdalla, A.; Sanga, H.; Mays, J.; Prior, T.; Jurgielewicz, A.; Guiry, S.; Davtyan, K. A Prospective Validation Cohort Study of Baseline Plasma Cell-Free DNA (CfDNA) as a Prognostic Biomarker in Newly Diagnosed Gli-oblastoma (GBM). Am. Soc. Clin. Oncol. 2020, 38, 2508. [Google Scholar] [CrossRef]
- Bagley, S.J.; Till, J.; Abdalla, A.; Sangha, H.K.; Yee, S.S.; Freedman, J.; Black, T.A.; Hussain, J.; Binder, Z.A.; Brem, S. Associa-tion of Plasma Cell-Free DNA with Survival in Patients with IDH Wild-Type Glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab011. [Google Scholar] [CrossRef]
- Miller, A.; Shah, R.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nat. Cell Biol. 2019, 565, 654–658. [Google Scholar] [CrossRef]
- Lu, V.M.; O’Connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neuro-Oncol. 2020, 148, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Aibaidula, A.; Chan, A.K.-Y.; Shi, Z.; Li, Y.; Zhang, R.; Yang, R.; Li, K.K.-W.; Chung, N.Y.-F.; Yao, Y.; Zhou, L.; et al. Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro-Oncology 2017, 19, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, R.; Song, C.; Xiang, Y.; Liu, Y. Prognostic and clinicopathological significance of long non-coding RNA in glioma. Neurosurg. Rev. 2018, 43, 1–8. [Google Scholar] [CrossRef]
- Iwamoto, F.M.; Hottinger, A.F.; Karimi, S.; Riedel, E.; Dantis, J.; Jahdi, M.; Panageas, K.S.; Lassman, A.B.; Abrey, L.E.; Fleisher, M.; et al. Serum YKL-40 is a marker of prognosis and disease status in high-grade gliomas. Neuro-Oncology 2011, 13, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; He, M.; Zhang, J.; Xu, J. High expression of microRNA 221 is a poor predictor for glioma. Medicine 2020, 99, e23163. [Google Scholar] [CrossRef]
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 Family as Novel Descriptive and Prognostic Biomarkers for Glioma. Mol. Neurobiol. 2015, 53, 1452–1460. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Xue, Q.; Wang, J.; Zhao, L.; Han, K.; Zhang, D.; Hou, L. Prognostic Significance of MicroRNAs in Glioma: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2019, 2019, 4015969. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, X.; Liu, Z.; Huang, X.; Li, X.; Cheng, K.; Jiang, X. Prognostic role of microRNA-155 expression in gliomas: A meta-analysis. Clin. Neurol. Neurosurg. 2018, 176, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Mu, J.; Liu, X.; Peng, X.; Zhong, F.; Yuan, W.; Deng, F.; Peng, X.; Peng, S.; Zeng, X. Prognostic value of miR-21 in gliomas: Comprehensive study based on meta-analysis and TCGA dataset validation. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, J.; Xiang, Q.; Liang, Y.; Zhao, N.; Zhang, Z.; Liu, Q.; Cui, Y. Prognostic role of microRNA-21 expression in gliomas: A meta-analysis. J. Neuro-Oncol. 2016, 130, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, J.; He, M.; Xu, J. Prognostic Role of MicroRNA 222 in Patients with Glioma: A Meta-analysis. BioMed Res. Int. 2020, 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Mancini, A.; Xavier-Magalhães, A.; Woods, W.S.; Nguyen, K.-T.; Amen, A.M.; Hayes, J.L.; Fellmann, C.; Gapinske, M.; McKinney, A.M.; Hong, C.; et al. Disruption of the β1L Isoform of GABP Reverses Glioblastoma Replicative Immortality in a TERT Promoter Mutation-Dependent Manner. Cancer Cell 2018, 34, 513–528. [Google Scholar] [CrossRef]
- Arita, H.; Narita, Y.; Fukushima, S.; Tateishi, K.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Collins, V.P.; Kawahara, N.; et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013, 126, 267–276. [Google Scholar] [CrossRef]
- Mosrati, M.A.; Malmström, A.; Lysiak, M.; Krysztofiak, A.; Hallbeck, M.; Milos, P.; Hallbeck, A.-L.; Bratthäll, C.; Strandéus, M.; Stenmark-Askmalm, M.; et al. TERT Promoter Mutations and Polymorphisms as Prognostic Factors in Primary Glioblastoma. Oncotarget 2015, 6, 16663–16673. [Google Scholar] [CrossRef]
- Shu, C.; Wang, Q.; Yan, X.; Wang, J. The TERT promoter mutation status and MGMT promoter methylation status, combined with dichotomized MRI-derived and clinical features, predict adult primary glioblastoma survival. Cancer Med. 2018, 7, 3704–3712. [Google Scholar] [CrossRef]
- Simon, M.; Hosen, I.; Gousias, K.; Rachakonda, S.; Heidenreich, B.; Gessi, M.; Schramm, J.; Hemminki, K.; Waha, A.; Kumar, R. TERT promoter mutations: A novel independent prognostic factor in primary glioblastomas. Neuro-Oncology 2014, 17, 45–52. [Google Scholar] [CrossRef]
- Labussiere, M.; Idbaih, A.; Wang, X.W.; Marie, Y.; Boisselier, B.; Falet, C.; Paris, S.; Laffaire, J.; Carpentier, C.; Criniere, E.; et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology 2010, 74, 1886–1890. [Google Scholar] [CrossRef]
- Spiegl-Kreinecker, S.; Lötsch, D.; Ghanim, B.; Pirker, C.; Mohr, T.; Laaber, M.; Weis, S.; Olschowski, A.; Webersinke, G.; Pichler, J.; et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: Interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro-Oncology 2015, 17, 1231–1240. [Google Scholar] [CrossRef]
- Yuan, Y.; Qi, C.; Maling, G.; Xiang, W.; Yanhui, L.; Ruofei, L.; Yunhe, M.; Jiewen, L.; Qing, M. TERT mutation in glioma: Frequency, prognosis and risk. J. Clin. Neurosci. 2016, 26, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Appay, R.; Dehais, C.; Maurage, C.-A.; Alentorn, A.; Carpentier, C.; Colin, C.; Ducray, F.; Escande, F.; Idbaih, A.; Kamoun, A.; et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro-Oncology 2019, 21, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Rudra, S.; Campian, J.L.; Dahiya, S.; Dunn, G.P.; Johanns, T.; Goldstein, M.; Kim, A.H.; Huang, J. Prognostic impact of CDKN2A/B deletion, TERT mutation, and EGFR amplification on histological and molecular IDH-wildtype glioblastoma. Neuro-Oncol. Adv. 2020, 2, vdaa126. [Google Scholar] [CrossRef] [PubMed]
- Manuel, J.M.; Ghosh, D.; Narasinga Rao, K.V.L.; Sibin, M.K.; Venkatesh, H.N.; Ch, L.; Chetan, G.K. Role of Concurrent Methylation Pattern of MGMT, TP53 and CDKN2A Genes in the Prognosis of High Grade Glioma. J. Carcinog. Mutagen. 2016, 7, 1. [Google Scholar] [CrossRef]
- Shirahata, M.; Ono, T.; Stichel, D.; Schrimpf, D.; Reuss, D.E.; Sahm, F.; Koelsche, C.; Wefers, A.; Reinhardt, A.; Huang, K.; et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018, 136, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, X.; Xu, L.; Zhang, J.; Cao, H. Analysis of the EGFR Amplification and CDKN2A Deletion Regulated Transcriptomic Signatures Reveals the Prognostic Significance of SPATS2L in Patients with Glioma. Front. Oncol. 2021, 11, 713. [Google Scholar] [CrossRef]
- Wu, F.; Li, G.; Liu, H.; Zhao, Z.; Chai, R.; Liu, Y.; Jiang, H.; Zhai, Y.; Feng, Y.; Li, R.; et al. Molecular subtyping reveals immune alterations in IDH wild-type lower-grade diffuse glioma. J. Pathol. 2020, 251, 272–283. [Google Scholar] [CrossRef]
- Yang, R.R.; Shi, Z.; Zhang, Z.; Chan, A.K.; Aibaidula, A.; Wang, W.; Kwan, J.S.H.; Poon, W.S.; Chen, H.; Li, W.; et al. IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2019, 30, 541–553. [Google Scholar] [CrossRef]
- Ghasimi, S.; Wibom, C.; Dahlin, A.; Brännström, T.; Golovleva, I.; Andersson, U.; Melin, B. Genetic risk variants in the CDKN2A/B, RTEL1 and EGFR genes are associated with somatic biomarkers in glioma. J. Neuro-Oncol. 2016, 127, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Alentorn, A.; Dehais, C.; Ducray, F.; Carpentier, C.; Mokhtari, K.; Figarella-Branger, D.; Chinot, O.; Cohen-Moyal, E.; Ramirez, C.; Loiseau, H.; et al. Allelic loss of 9p21.3 is a prognostic factor in 1p/19q codeleted anaplastic gliomas. Neurology 2015, 85, 1325–1331. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, M.C.; Born, N. Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. J. Neuro-Oncol. 2015, 125, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.B.; Blair, H.; Ballman, K.V.; Giannini, C.; Arusell, R.M.; Law, M.; Flynn, H.; Passe, S.; Felten, S.; Brown, P.D.; et al. A t(1;19)(q10;p10) Mediates the Combined Deletions of 1p and 19q and Predicts a Better Prognosis of Patients with Oligodendroglioma. Cancer Res. 2006, 66, 9852–9861. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.W.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERTPromoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Jiang, H.; Ren, X.; Cui, X.; Wang, J.; Jia, W.; Zhou, Z.; Lin, S. 1p/19q codeletion and IDH1/2 mutation identified a subtype of anaplastic oligoastrocytomas with prognosis as favorable as anaplastic oligodendrogliomas. Neuro-Oncology 2013, 15, 775–782. [Google Scholar] [CrossRef]
- Reuss, D.E.; Sahm, F.; Schrimpf, D.; Wiestler, B.; Capper, D.; Koelsche, C.; Schweizer, L.; Korshunov, A.; Jones, D.T.W.; Hovestadt, V.; et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2014, 129, 133–146. [Google Scholar] [CrossRef]
- Huse, J.T.; Diamond, E.; Wang, L.; Rosenblum, M.K. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: A true “oligoastrocytoma”? Acta Neuropathol. 2014, 129, 151–153. [Google Scholar] [CrossRef]
- Hu, X.; Martinez-Ledesma, E.; Zheng, S.; Kim, H.; Barthel, F.; Jiang, T.; Hess, K.R.; Verhaak, R.G. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro-Oncology 2017, 19, 786–795. [Google Scholar] [CrossRef]
- Kaloshi, G.; Benouaich-Amiel, A.; Diakite, F.; Taillibert, S.; Lejeune, J.; Laigle-Donadey, F.; Renard, M.-A.; Iraqi, W.; Idbaih, A.; Paris, S.; et al. Temozolomide for low-grade gliomas: Predictive impact of 1p/19q loss on response and outcome. Neurology 2007, 68, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Vogazianou, A.P.; Chan, R.; Bäcklund, L.M.; Pearson, D.M.; Liu, L.; Langford, C.F.; Gregory, S.G.; Collins, V.P.; Ichimura, K. Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses. Neuro-Oncology 2010, 12, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Stupp, R.; Hegi, M.; Bent, M.V.D.; Tonn, J.C.; Sanson, M.; Wick, W.; Reifenberger, G. Personalized care in neuro-oncology coming of age: Why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro-Oncology 2012, 14, iv100–iv108. [Google Scholar] [CrossRef] [PubMed]
- Cairncross, G.; Berkey, B.; Shaw, E.; Jenkins, R.; Scheithauer, B.; Brachman, D.; Buckner, J.; Fink, K.; Souhami, L.; Laperierre, N.; et al. Phase III Trial of Chemotherapy Plus Radiotherapy Compared with Radiotherapy Alone for Pure and Mixed Anaplastic Oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J. Clin. Oncol. 2006, 24, 2707–2714. [Google Scholar] [CrossRef]
- Erdem-Eraslan, L.; Gravendeel, L.A.; de Rooi, J.; Eilers, P.H.; Idbaih, A.; Spliet, W.G.; Dunnen, W.F.D.; Teepen, J.L.; Wesseling, P.; Smitt, P.A.S.; et al. Intrinsic Molecular Subtypes of Glioma Are Prognostic and Predict Benefit from Adjuvant Procarbazine, Lomustine, and Vincristine Chemotherapy in Combination with Other Prognostic Factors in Anaplastic Oligodendroglial Brain Tumors: A Report From EORTC Study 26951. J. Clin. Oncol. 2013, 31, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef]
- Balesaria, S.; Brock, C.; Bower, M.; Clark, J.; Nicholson, S.K.; Lewis, P.; De Sanctis, S.; Evans, H.; Peterson, D.; Mendoza, N.; et al. Loss of chromosome 10 is an independent prognostic factor in high-grade gliomas. Br. J. Cancer 1999, 81, 1371–1377. [Google Scholar] [CrossRef]
- Roth, J.J.; Fierst, T.M.; Waanders, A.J.; Yimei, L.; Biegel, J.A.; Santi, M. Whole Chromosome 7 Gain Predicts Higher Risk of Recurrence in Pediatric Pilocytic Astrocytomas Independently from KIAA1549-BRAF Fusion Status. J. Neuropathol. Exp. Neurol. 2016, 75, 306–315. [Google Scholar] [CrossRef][Green Version]
- Richard, S.; Tachon, G.; Milin, S.; Wager, M.; Karayan-Tapon, L. Dual MGMT inactivation by promoter hypermethylation and loss of the long arm of chromosome 10 in glioblastoma. Cancer Med. 2020, 9, 6344–6353. [Google Scholar] [CrossRef]
- Zhou, Y.; Ness, S.A. Myb Proteins: Angels and Demons in Normal and Transformed Cells. Front. Biosci. J. Virtual Libr. 2011, 16, 1109–1131. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-H.; Harreld, J.H.; Tinkle, C.L.; Moreira, D.C.; Li, X.; Acharya, S.; Qaddoumi, I.; Ellison, D.W. A single-center study of the clinicopathologic correlates of gliomas with a MYB or MYBL1 alteration. Acta Neuropathol. 2019, 138, 1091–1092. [Google Scholar] [CrossRef]
- Ellison, D.W.; Hawkins, C.; Jones, D.T.W.; Onar-Thomas, A.; Pfister, S.M.; Reifenberger, G.; Louis, D.N. cIMPACT-NOW update 4: Diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019, 137, 683–687. [Google Scholar] [CrossRef]
- Wood, M.D.; Tihan, T.; Perry, A.; Chacko, G.; Turner, C.; Pu, C.; Payne, C.; Yu, A.; Bannykh, S.I.; Solomon, D.A. Multimodal molecular analysis of astroblastoma enables reclassification of most cases into more specific molecular entities. Brain Pathol. 2017, 28, 192–202. [Google Scholar] [CrossRef]
- Saini, M.; Jha, A.N.; Tangri, R.; Qudratullah, M.; Ali, S. MN1 overexpression with varying tumor grade is a promising predictor of survival of glioma patients. Hum. Mol. Genet. 2020, 29, 3532–3545. [Google Scholar] [CrossRef]
- Mhatre, R.; Sugur, H.S.; Nandeesh, B.N.; Chickabasaviah, Y.; Saini, J.; Santosh, V. MN1 rearrangement in astroblastoma: Study of eight cases and review of literature. Brain Tumor Pathol. 2019, 36, 112–120. [Google Scholar] [CrossRef]
- Lehman, N.L.; Usubalieva, A.; Lin, T.; Allen, S.J.; Tran, Q.T.; Mobley, B.C.; McLendon, R.E.; Schniederjan, M.J.; Georgescu, M.-M.; Couce, M.; et al. Genomic analysis demonstrates that histologically-defined astroblastomas are molecularly heterogeneous and that tumors with MN1 rearrangement exhibit the most favorable prognosis. Acta Neuropathol. Commun. 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Santarpia, L.; Lippman, S.L.; El-Naggar, A.K. Targeting the MAPK–RAS–RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Stichel, D.; Schrimpf, D.; Sahm, F.; Korshunov, A.; Reuss, D.E.; Koelsche, C.; Huang, K.; Wefers, A.K.; Hovestadt, V.; et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018, 136, 273–291. [Google Scholar] [CrossRef]
- Jones, D.T.W.; The International Cancer Genome Consortium PedBrain Tumor Project; Hutter, B.; Jaeger, N.; Korshunov, A.; Kool, M.; Warnatz, H.-J.; Zichner, T.; Lambert, S.R.; Ryzhova, M.; et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet. 2013, 45, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.W.; Gronych, J.; Lichter, P.; Witt, O.; Pfister, S.M. MAPK pathway activation in pilocytic astrocytoma. Experientia 2011, 69, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.P.; Scapulatempo-Neto, C.; Carloni, A.C.; Paulino, A.; Sheren, J.; Aisner, D.L.; Musselwhite, E.; Clara, C.; Machado, H.R.; Oliveira, R.S.; et al. KIAA1549: BRAF Gene Fusion and FGFR1 Hotspot Mutations Are Prognostic Factors in Pilo-cytic Astrocytomas. J. Neuropathol. Exp. Neurol. 2015, 74, 743–754. [Google Scholar] [CrossRef]
- Arteaga, C.L.; Engelman, J.A. ERBB Receptors: From Oncogene Discovery to Basic Science to Mechanism-Based Cancer Therapeutics. Cancer Cell 2014, 25, 282–303. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440–33450. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Schmidt, M.H.H. EGFR and EGFRvIII Promote Angiogenesis and Cell Invasion in Glioblastoma: Combination Therapies for an Effective Treatment. Int. J. Mol. Sci. 2017, 18, 1295. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.K.; Kaye, A.H.; Luwor, R.B. The EGFRvIII variant in glioblastoma multiforme. J. Clin. Neurosci. 2009, 16, 748–754. [Google Scholar] [CrossRef]
- Eskilsson, E.; Rosland, G.V.; Talasila, K.M.; Knappskog, S.; Keunen, O.; Sottoriva, A.; Foerster, S.; Solecki, G.; Taxt, T.; Jirik, R.; et al. EGFRvIII mutations can emerge as late and heterogenous events in glioblastoma development and promote angiogenesis through Src activation. Neuro-Oncology 2016, 18, 1644–1655. [Google Scholar] [CrossRef]
- Bale, T.A.; Jordan, J.T.; Rapalino, O.; Ramamurthy, N.; Jessop, N.; DeWitt, J.C.; Nardi, V.; Alvarez, M.M.-L.; Frosch, M.; Batchelor, T.T.; et al. Financially effective test algorithm to identify an aggressive, EGFR-amplified variant of IDH-wildtype, lower-grade diffuse glioma. Neuro-Oncology 2018, 21, 596–605. [Google Scholar] [CrossRef]
- Labussière, M.; Boisselier, B.; Mokhtari, K.; Di Stefano, A.-L.; Rahimian, A.; Rossetto, M.; Ciccarino, P.; Saulnier, O.; Paterra, R.; Marie, Y.; et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology 2014, 83, 1200–1206. [Google Scholar] [CrossRef]
- Hao, Z.; Guo, D. EGFR mutation: Novel prognostic factor associated with immune infiltration in lower-grade glioma; an exploratory study. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Shinojima, N.; Tada, K.; Shiraishi, S.; Kamiryo, T.; Kochi, M.; Nakamura, H.; Makino, K.; Saya, H.; Hirano, H.; Kuratsu, J.-I.; et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003, 63, 6962–6970. [Google Scholar]
- Lee, K.; Koo, H.; Kim, Y.; Kim, D.; Son, E.; Yang, H.; Lim, Y.; Hur, M.; Lee, H.; Choi, S.; et al. Therapeutic Efficacy of GC1118, a Novel Anti-Egfr Antibody, against Glioblastoma with High Egfr Amplification in Patient-Derived Xenografts. Cancers 2020, 12, 3210. [Google Scholar] [CrossRef] [PubMed]
- Nadeem Abbas, M.; Kausar, S.; Wang, F.; Zhao, Y.; Cui, H. Cui Advances in Targeting the Epidermal Growth Factor Receptor Pathway by Synthetic Products and Its Regulation by Epigenetic Modulators as a Therapy for Glioblastoma. Cells 2019, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Yang, Y.; Wang, Q.; Bergholz, J.S.; Jiang, T.; Roberts, T.M.; Gray, N.S.; Zhao, J.J. Targeting EGFR in Glioblastoma with a Novel Brain-Penetrant Small Molecule EGFR-TKI. BioRxiv 2021, 2021, 426030. [Google Scholar] [CrossRef]
- Zając, A.; Sumorek-Wiadro, J.; Langner, E.; Wertel, I.; Maciejczyk, A.; Pawlikowska-Pawlęga, B.; Pawelec, J.; Wasiak, M.; Hułas-Stasiak, M.; Bądziul, D.; et al. Involvement of PI3K Pathway in Glioma Cell Resistance to Temozolomide Treatment. Int. J. Mol. Sci. 2021, 22, 5155. [Google Scholar] [CrossRef] [PubMed]
- Duque, M.B.; Pinheiro, K.D.V.; Thomaz, A.; Da Silva, C.A.; Freire, N.H.; Brunetto, A.T.; Schwartsmann, G.; Jaeger, M.; de Farias, C.; Roesler, R. Combined Inhibition of HDAC and EGFR Reduces Viability and Proliferation and Enhances STAT3 mRNA Expression in Glioblastoma Cells. J. Mol. Neurosci. 2019, 68, 49–57. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Tong, L.; Luo, Y.; Su, M.; Zang, Y.; Li, J.; Lu, W.; Chen, Y. The discovery of colchicine-SAHA hybrids as a new class of antitumor agents. Bioorgan. Med. Chem. 2013, 21, 3240–3244. [Google Scholar] [CrossRef]
- Zhang, I.; Beus, M.; Stochaj, U.; Le, P.U.; Zorc, B.; Rajić, Z.; Petrecca, K.; Maysinger, D. Inhibition of glioblastoma cell proliferation, invasion, and mechanism of action of a novel hydroxamic acid hybrid molecule. Cell Death Discov. 2018, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Dolan, M.E.; Moschel, R.C. Structure, Function, and Inhibition of O6-Alkylguanine-DNA Alkyltransferase. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Moldave, K., Eds.; Academic Press: Cambridge, MA, USA, 1995; Volume 51, pp. 167–223. [Google Scholar]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.; Gilbert, M.R. Correlation of O6-Methylguanine Methyltransferase (MGMT) Promoter Methylation with Clinical Outcomes in Glioblastoma and Clinical Strategies to Modulate MGMT Activity. J. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, X.-Q.; Zhou, B.; Zhang, L. The prognostic value of MGMT promoter methylation in Glioblastoma multiforme: A meta-analysis. Fam. Cancer 2013, 12, 449–458. [Google Scholar] [CrossRef]
- Yin, A.-A.; Zhang, L.-H.; Cheng, J.-X.; Dong, Y.; Liu, B.-L.; Han, N.; Zhang, X. The Predictive but Not Prognostic Value of MGMT Promoter Methylation Status in Elderly Glioblastoma Patients: A Meta-Analysis. PLoS ONE 2014, 9, e85102. [Google Scholar] [CrossRef] [PubMed]
- Thon, N.; Kreth, S.; Kreth, F.W. Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. OncoTargets Ther. 2013, 6, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Goodman, S.N.; Herman, J.G. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Hegi, M.E.; Hamou, M.-F.; de Tribolet, N.; Kros, J.M.; Mariani, L.; Mirimanoff, R.O.; Stupp, R. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 52, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Abacioglu, U.; Tavelin, B.; Lhermitte, B.; Hegi, M.; et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef]
- Vaubel, R.A.; Tian, S.; Remonde, D.A.; Schroeder, M.A.; Mladek, A.C.; Kitange, G.J.; Caron, A.; Kollmeyer, T.M.; Grove, R.; Peng, S.; et al. Genomic and Phenotypic Characterization of a Broad Panel of Patient-Derived Xenografts Reflects the Diversity of Glioblastoma. Clin. Cancer Res. 2019, 26, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Roszkowski, K.; Furtak, J.; Zurawski, B.; Szylberg, T.; Lewandowska, M.A. Potential Role of Methylation Marker in Glioma Supporting Clinical Decisions. Int. J. Mol. Sci. 2016, 17, 1876. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.; Costa, B.; Caeiro, C.; Guimarães, I.; Martinho, O.; Jaraquemada, T.; Augusto, I.; Castro, L.; Osório, L.; Linhares, P.; et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: A Portuguese multicentre study. Oncol. Rep. 2010, 23, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro-Oncology 2018, 21, 167–178. [Google Scholar] [CrossRef]
- Radke, J.; Koch, A.; Pritsch, F.; Schumann, E.; Misch, M.; Hempt, C.; Lenz, K.; Löbel, F.; Paschereit, F.; Heppner, F.; et al. Predictive MGMT status in a homogeneous cohort of IDH wildtype glioblastoma patients. Acta Neuropathol. Commun. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Chai, E.A.R.; Li, G.; Liu, Y.; Zhang, K.; Zhao, Z.; Wu, F.; Chang, Y.; Pang, B.; Li, J.; Li, Y.; et al. Predictive value of MGMT promoter methylation on the survival of TMZ treated IDH-mutant glioblastoma. Cancer Biol. Med. 2021, 18, 271–282. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef]
- Reifenberger, G.; Hentschel, B.; Felsberg, J.; Schackert, G.; Simon, M.; Schnell, O.; Westphal, M.; Wick, W.; Pietsch, T.; Loeffler, M.; et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int. J. Cancer 2011, 131, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Lin, B.; Sibenaller, Z.; Ryken, T.; Lee, H.; Yoon, J.-G.; Rostad, S.; Foltz, G. Comprehensive Analysis of MGMT Promoter Methylation: Correlation with MGMT Expression and Clinical Response in GBM. PLoS ONE 2011, 6, e16146. [Google Scholar] [CrossRef]
- Boots-Sprenger, S.H.; Sijben, A.; Rijntjes, J.; Tops, B.B.; Idema, A.J.; Rivera, A.L.; Bleeker, F.E.; Gijtenbeek, A.M.; Diefes, K.; Heathcock, L.; et al. Significance of Complete 1p/19q Co-Deletion, IDH1 Mutation and MGMT Promoter Methylation in Gli-omas: Use with Caution. Mod. Pathol. 2013, 26, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Lie, A.; Li, T.; Chowdhury, R.; Liu, F.; Ozer, B.; Wei, B.; Green, R.M.; Ellingson, B.M.; Wang, H.-J.; et al. HumanTERTpromoter mutation enables survival advantage from MGMT promoter methylation inIDH1wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro-Oncol. 2016, 19, 394–404. [Google Scholar] [CrossRef]
- Dahlrot, R.; Larsen, P.; Boldt, H.B.; Kreutzfeldt, M.S.; Hansen, S.; Hjelmborg, J.B.; Kristensen, B.W. Posttreatment Effect of MGMT Methylation Level on Glioblastoma Survival. J. Neuropathol. Exp. Neurol. 2019, 78, 633–640. [Google Scholar] [CrossRef]
- Von Bueren, A.O.; Bacolod, M.D.; Hagel, C.; Heinimann, K.; Fedier, A.; Kordes, U.; Pietsch, T.; Koster, J.; Grotzer, M.; Friedman, H.S.; et al. Mismatch repair deficiency: A temozolomide resistance factor in medulloblastoma cell lines that is uncommon in primary medulloblastoma tumours. Br. J. Cancer 2012, 107, 1399–1408. [Google Scholar] [CrossRef]
- Jiapaer, S.; Furuta, T.; Tanaka, S.; Kitabayashi, T.; Nakada, M. Potential Strategies Overcoming the Temozolomide Resistance for Glioblastoma. Neurol. Med. Chir. 2018, 58, 405–421. [Google Scholar] [CrossRef]
- Suwala, A.K.; Stichel, D.; Schrimpf, D.; Kloor, M.; Wefers, A.K.; Reinhardt, A.; Maas, S.L.N.; Kratz, C.P.; Schweizer, L.; Hasselblatt, M.; et al. Primary mismatch repair deficient IDH-mutant astrocytoma (PMMRDIA) is a distinct type with a poor prognosis. Acta Neuropathol. 2020, 141, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Caccese, M.; Ius, T.; Simonelli, M.; Fassan, M.; Cesselli, D.; DiPasquale, A.; Cavallin, F.; Padovan, M.; Salvalaggio, A.; Gardiman, M.P.; et al. Mismatch-Repair Protein Expression in High-Grade Gliomas: A Large Retrospective Multicenter Study. Int. J. Mol. Sci. 2020, 21, 6716. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yu, Y.; Grimmer, M.R.; Wahl, M.; Chang, S.M.; Costello, J.F. Temozolomide-associated hypermutation in gliomas. Neuro-Oncology 2018, 20, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Touat, M.; Li, Y.Y.; Boynton, A.N.; Spurr, L.F.; Iorgulescu, J.B.; Bohrson, C.L.; Cortes-Ciriano, I.; Birzu, C.; Geduldig, J.E.; Pelton, K.; et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 2020, 580, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Lemery, S.; Keegan, P.; Pazdur, R. First FDA Approval Agnostic of Cancer Site—When a Biomarker Defines the Indication. Available online: https://www.nejm.org/doi/10.1056/NEJMp1709968 (accessed on 20 July 2021).
- McGranahan, N.; Furness, A.J.S.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.; Hiley, C.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Daniel, P.; Sabri, S.; Chaddad, A.; Meehan, B.; Jean-Claude, B.; Rak, J.; Abdulkarim, B.S. Temozolomide Induced Hypermutation in Glioma: Evolutionary Mechanisms and Therapeutic Opportunities. Front. Oncol. 2019, 9, 41. [Google Scholar] [CrossRef]
- Bark, J.M.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer 2019, 122, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, D.; Li, W.; Xiang, X.; Zhao, J.; Yu, B.; Wang, C.; He, Z.; Zhu, L.; Yang, Y. Evaluation of serum extracellular vesicles as noninvasive diagnostic markers of glioma. Theranostics 2019, 9, 5347–5358. [Google Scholar] [CrossRef]
- Engelborghs, S.; Niemantsverdriet, E.; Struyfs, H.; Blennow, K.; Brouns, R.; Comabella, M.; Dujmovic, I.; van der Flier, W.; Froelich, L.; Galimberti, D.; et al. Consensus guidelines for lumbar puncture in patients with neurological diseases. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2017, 8, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Mair, R.; Mouliere, F.; Smith, C.G.; Chandrananda, D.; Gale, D.; Marass, F.; Tsui, D.W.; Massie, C.E.; Wright, A.J.; Watts, C. Measurement of Plasma Cell-Free Mitochondrial Tumor DNA Improves Detection of Glioblastoma in Patient-Derived Ortho-topic Xenograft Models. Cancer Res. 2019, 79, 220–230. [Google Scholar] [CrossRef]
- Wan, J.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.C.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Cordova, C.; Syeda, M.M.; Corless, B.; Wiggins, J.M.; Patel, A.; Kurz, S.C.; Delara, M.; Sawaged, Z.; Utate, M.; Placantonakis, D. Abstract A65: Longitudinal Detection of TERT-Mutant Plasma Cell-Free Circulating Tumor DNA in Newly Diagnosed Glioblastoma Patients; AACR: Philadelphia, PA, USA, 2020. [Google Scholar]
- Piccioni, D.E.; Achrol, A.S.; Kiedrowski, L.A.; Banks, K.; Boucher, N.; Barkhoudarian, G.; Kelly, D.F.; Juarez, T.; Lanman, R.B.; Raymond, V.M.; et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol. 2019, 8, CNS34. [Google Scholar] [CrossRef]
- Liang, J.; Zhao, W.; Lu, C.; Liu, D.; Li, P.; Ye, X.; Zhao, Y.; Zhang, J.; Yang, D. Next-Generation Sequencing Analysis of ctDNA for the Detection of Glioma and Metastatic Brain Tumors in Adults. Front. Neurol. 2020, 11, 544. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, C.; Li, M.; Shen, Y.; Feng, S.; Liu, J.; Li, F.; Hou, L.; Chen, Z.; Jiang, J.; et al. Applications of cerebrospinal fluid circulating tumor DNA in the diagnosis of gliomas. Jpn. J. Clin. Oncol. 2020, 50, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; He, Z.-Q.; Lin, F.-H.; Chen, Z.-H.; Yang, S.-Y.; Duan, H.; Jiang, X.-B.; Al-Nahari, F.; Zhang, X.-H.; Wang, J.-H.; et al. Assessment of ctDNA in CSF may be a more rapid means of assessing surgical outcomes than plasma ctDNA in glioblastoma. Mol. Cell. Probes 2019, 46, 101411. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, J.; Xu, J.; Zheng, C.; Li, X.; Du, J. The analysis of microRNA-34 family expression in human cancer studies comparing cancer tissues with corresponding pericarcinous tissues. Gene 2015, 554, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nørøxe, D.S.; Østrup, O.; Yde, C.W.; Ahlborn, L.B.; Nielsen, F.C.; Michaelsen, S.R.; Larsen, V.A.; Skjøth-Rasmussen, J.; Bren-num, J.; Hamerlik, P. Cell-Free DNA in Newly Diagnosed Patients with Glioblastoma—A Clinical Prospective Feasibility Study. Oncotarget 2019, 10, 4397. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 1–10. [Google Scholar] [CrossRef]
- Heredia-Soto, V.; Rodríguez-Salas, N.; Feliu, J. Liquid Biopsy in Pancreatic Cancer: Are We Ready to Apply It in the Clinical Practice? Cancers 2021, 13, 1986. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, eaaz8084. [Google Scholar] [CrossRef]
- Bauml, J.; Levy, B. Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clin. Cancer Res. 2018, 24, 4352–4354. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kamińska, K.; Nalejska, E.; Kubiak, M.; Wojtysiak, J.; Żołna, Ł.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Epigenetic Biomarkers in Oncology. Mol. Diagn. Ther. 2018, 23, 83–95. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zöllner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro-Oncology 2011, 14, 29–33. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, J.; Quan, J.; Liu, W.; Tan, H.; Li, W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci. 2018, 109, 2651–2659. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients: Glioblastoma-Associated MiRNA Profile in Peripheral Blood. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef]
- Wang, J.; Che, F.; Zhang, J. Cell-free microRNAs as non-invasive biomarkers in glioma: A diagnostic meta-analysis. Int. J. Biol. Markers 2019, 34, 232–242. [Google Scholar] [CrossRef] [PubMed]
- ParvizHamidi, M.; Haddad, G.; Ostadrahimi, S.; Ostadrahimi, N.; Sadeghi, S.; Fayaz, S.; Fard-Esfahani, P. Circulating MiR-26a and MiR-21 as Biomarkers for Glioblastoma Multiform. Biotechnol. Appl. Biochem. 2019, 66, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, D.; Lv, T.; Li, G.; Qu, S. Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol. Neurobiol. 2014, 53, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Phillips, S.; Kaimal, V.; Mao, Y.; Hua, W.; Yang, I.; Fu, C.-C.; Nolan, J.; et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J. Neuro-Oncol. 2015, 123, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncology 2012, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.J.A.; Balaj, L.; Hulleman, E.; Van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.; Vandertop, W.P.; et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M.; et al. Human Glioma Growth Is Controlled by MicroRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Luo, Q.; Han, Y.; Huang, D.; Tang, Q.; Wu, L. MiR-223/PAX6 Axis Regulates Glioblastoma Stem Cell Proliferation and the Chemo Resistance to TMZ via Regulating PI3K/Akt Pathway. J. Cell. Biochem. 2017, 118, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, S.; Feng, K.; Wu, F.; Wan, Y.; Wang, Z.; Zhang, J.; Wang, Y.; Yan, W.; Fu, Z.; et al. MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int. J. Oncol. 2011, 40, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Siegal, T.; Charbit, H.; Paldor, I.; Zelikovitch, B.; Canello, T.; Benis, A.; Wong, M.L.; Morokoff, A.; Kaye, A.H.; Lavon, I. Dynamics of circulating hypoxia-mediated miRNAs and tumor response in patients with high-grade glioma treated with bevacizumab. J. Neurosurg. 2016, 125, 1008–1015. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, J.; Hoadley, K.; Kushwaha, D.; Ramakrishnan, V.; Li, S.; Kang, C.; You, Y.; Jiang, C.; Song, S.W.; et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro-Oncology 2012, 14, 712–719. [Google Scholar] [CrossRef]
- Shi, L.; Chen, J.; Yang, J.; Pan, T.; Zhang, S.; Wang, Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010, 1352, 255–264. [Google Scholar] [CrossRef]
- Gwak, H.-S.; Kim, T.H.; Jo, G.H.; Kim, Y.-J.; Kwak, H.-J.; Kim, J.H.; Yin, J.; Yoo, H.; Lee, S.H.; Park, J.B. Silencing of MicroRNA-21 Confers Radio-Sensitivity through Inhibition of the PI3K/AKT Pathway and Enhancing Autophagy in Malignant Glioma Cell Lines. PLoS ONE 2012, 7, e47449. [Google Scholar] [CrossRef]
- Akers, J.C.; Hua, W.; Li, H.; Ramakrishnan, V.; Yang, Z.; Quan, K.; Zhu, W.; Li, J.; Figueroa, J.; Hirshman, B.R. A Cerebrospinal Fluid MicroRNA Signature as Biomarker for Glioblastoma. Oncotarget 2017, 8, 68769. [Google Scholar] [CrossRef]
- Buruiană, A.; Florian, Ș.I.; Florian, A.I.; Timiș, T.-L.; Mihu, C.M.; Miclăuș, M.; Oșan, S.; Hrapșa, I.; Cataniciu, R.C.; Far-caș, M. The Roles of MiRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950. [Google Scholar] [CrossRef]
- Conti, A.; Aguennouz, M.; La Torre, D.; Tomasello, C.; Cardali, S.M.; Angileri, F.; Maio, F.; Cama, A.; Germanò, A.; Vita, G.; et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J. Neuro-Oncol. 2009, 93, 325–332. [Google Scholar] [CrossRef]
- Møller, H.G.; Rasmussen, A.P.; Andersen, H.H.; Johnsen, K.B.; Henriksen, M.; Duroux, M. A Systematic Review of Mi-croRNA in Glioblastoma Multiforme: Micro-Modulators in the Mesenchymal Mode of Migration and Invasion. Mol. Neurobiol. 2013, 47, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Rolle, K.; Belter, A.; Barciszewska, A.-M.; Żywicki, M.; Michalak, M.; Nowak, S.; Naskręt-Barciszewska, M.Z.; Barciszewski, J. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol. Oncol. 2015, 9, 1324–1340. [Google Scholar] [CrossRef] [PubMed]
- Visani, M.; De Biase, D.; Marucci, G.; Cerasoli, S.; Nigrisoli, E.; Reggiani, M.L.B.; Albani, F.; Baruzzi, A.; Pession, A. The PERNO study group Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I–III. Mol. Oncol. 2013, 8, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wei, W.; Zhang, Z.; He, C.; Yang, R.; Zhang, J.; Wu, Z.; Huang, Q.; Jiang, Q. Identification of microRNAs associated with glioma diagnosis and prognosis. Oncotarget 2017, 8, 26394–26403. [Google Scholar] [CrossRef]
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. 2012, 31, 97. [Google Scholar] [CrossRef]
- Li, H.-Y.; Li, Y.-M.; Shi, X.-W.; Chen, H. Circulating microRNA-137 is a potential biomarker for human glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3599–3604. [Google Scholar]
- Garcia, C.M.; Toms, S.A. The Role of Circulating MicroRNA in Glioblastoma Liquid Biopsy. World Neurosurg. 2020, 138, 425–435. [Google Scholar] [CrossRef]
- Ciafrè, S.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.-G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.; Farace, M. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Patric, I.R.P.; Somasundaram, K. A Ten-MicroRNA Expression Signature Predicts Survival in Glioblastoma. PLoS ONE 2011, 6, e17438. [Google Scholar] [CrossRef]
- Møllersen, K.; Zortea, M.; Schopf, T.R.G.; Kirchesch, H.M.; Godtliebsen, F. Comparison of computer systems and ranking criteria for automatic melanoma detection in dermoscopic images. PLoS ONE 2017, 12, e0190112. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Han, C.; Wang, X.; She, L.; Zhang, H. miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int. J. Oncol. 2014, 45, 746–756. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, S.; Pu, J.K.S.; Tsang, A.C.O.; Lee, D.; Man, V.O.Y.; Lui, W.M.; Wong, T.S.; Leung, G.K.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, C.; Wu, M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 2018, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, J.; Wu, P.; Yang, W.; Ye, Q.; Chen, Q.; Jiang, C. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-κB pathway. J. Neuro-Oncol. 2018, 140, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, Q.; Li, Z.; Hu, Y.; Zhou, F.; Zhai, Z.; Yue, S.; Tian, H. LncRNA LINC00319 is associated with tumorigenesis and poor prognosis in glioma. Eur. J. Pharmacol. 2019, 861, 172556. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Q.; Ge, P.; Zeng, C.; Lin, F.; Wang, W.; Zhao, J. FAM225B Is a Prognostic lncRNA for Patients with Recurrent Glioblastoma. Dis. Markers 2020, 2020, 1–7. [Google Scholar] [CrossRef]
- Aslan, K.; Turco, V.; Blobner, J.; Sonner, J.K.; Liuzzi, A.R.; Núñez, N.G.; De Feo, D.; Kickingereder, P.V.; Fischer, M.; Green, E.; et al. Heterogeneity of response to immune checkpoint blockade in hypermutated experimental gliomas. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, Z.; Zhao, H.; Bao, S.; Cheng, L.; Sun, J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol. Neurobiol. 2017, 55, 1–14. [Google Scholar] [CrossRef]
- Luan, F.; Chen, W.; Chen, M.; Yan, J.; Chen, H.; Yu, H.; Liu, T.; Mo, L. An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio 2019, 9, 653–667. [Google Scholar] [CrossRef]
- Wu, C.; Su, J.; Long, W.; Qin, C.; Wang, X.; Xiao, K.; Li, Y.; Xiao, Q.; Wang, J.; Pan, Y.; et al. LINC00470 promotes tumour proliferation and invasion, and attenuates chemosensitivity through the LINC00470/miR-134/Myc/ABCC1 axis in glioma. J. Cell. Mol. Med. 2020, 24, 12094–12106. [Google Scholar] [CrossRef]
- Yang, J.-K.; Yang, J.-P.; Tong, J.; Jing, S.-Y.; Fan, B.; Wang, F.; Sun, G.-Z.; Jiao, B.-H. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. Neuro-Oncol. 2016, 131, 255–265. [Google Scholar] [CrossRef]
- Fan, Q.; Yang, L.; Zhang, X.; Peng, X.; Wei, S.; Su, D.; Zhai, Z.; Hua, X.; Li, H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2017, 414, 107–115. [Google Scholar] [CrossRef]
- Chae, Y.; Roh, J.; Kim, W. The Roles Played by Long Non-Coding RNAs in Glioma Resistance. Int. J. Mol. Sci. 2021, 22, 6834. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Zijl, S.; Reijneveld, J.C.; Wesseling, P.; Wurdinger, T. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol. 2015, 129, 849–865. [Google Scholar] [CrossRef]
- Hormigo, A.; Gu, B.; Karimi, S.; Riedel, E.; Panageas, K.S.; Edgar, M.A.; Tanwar, M.K.; Rao, J.S.; Fleisher, M.; DeAngelis, L.; et al. YKL-40 and Matrix Metalloproteinase-9 as Potential Serum Biomarkers for Patients with High-Grade Gliomas. Clin. Cancer Res. 2006, 12, 5698–5704. [Google Scholar] [CrossRef]
- Vaitkiene, P.; Urbanaviciute, R.; Grigas, P.; Steponaitis, G.; Tamasauskas, A.; Skiriutė, D. Identification of Astrocytoma Blood Serum Protein Profile. Cells 2019, 9, 16. [Google Scholar] [CrossRef]
- Tabouret, E.; Boudouresque, F.; Barrie, M.; Matta, M.; Boucard, C.; Loundou, A.; Carpentier, A.; Sanson, M.; Metellus, P.; Figarella-Branger, M.; et al. Association of matrix metalloproteinase 2 plasma level with response and survival in patients treated with bevacizumab for recurrent high-grade glioma. Neuro-Oncology 2013, 16, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, P.; Chowdhary, S.; Potthast, L.; Prabhu, A.; Tsai, Y.-Y.; Sarcar, B.; Kahali, S.; Brem, S.; Yu, H.M.; Rojiani, A.; et al. Phase I trial of vorinostat combined with bevacizumab and CPT-11 in recurrent glioblastoma. Neuro-Oncology 2011, 14, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Basu, B.; Ghosh, M.K. Extracellular Vesicles in Glioma: From Diagnosis to Therapy. BioEssays 2019, 41, e1800245. [Google Scholar] [CrossRef]
- Quezada, C.; Torres, Á.; Niechi, I.; Uribe, D.; Contreras-Duarte, S.; Toledo, F.; Martín, R.S.; Gutiérrez, J.; Sobrevia, L. Role of extracellular vesicles in glioma progression. Mol. Asp. Med. 2018, 60, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Westphal, M.; Lamszus, K. Circulating biomarkers for gliomas. Nat. Rev. Neurol. 2015, 11, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Z.; Mashimo, T.; Shen, B.; Nyagilo, J.; Wang, H.; Wang, Y.; Liu, Z.; Mulgaonkar, A.; Hu, X.-L.; et al. Gliomas Interact with Non-glioma Brain Cells via Extracellular Vesicles. Cell Rep. 2020, 30, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Tom, M.C.; Cahill, D.P.; Buckner, J.C.; Dietrich, J.; Parsons, M.W.; Yu, J.S. Management for Different Glioma Subtypes: Are All Low-Grade Gliomas Created Equal? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 133–145. [Google Scholar] [CrossRef] [PubMed]
- André-Grégoire, G.; Bidère, N.; Gavard, J. Temozolomide affects Extracellular Vesicles Released by Glioblastoma Cells. Biochimie 2018, 155, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Azam, Z.; Quillien, V.; Wang, G.; To, S.-S.T. The potential diagnostic and prognostic role of extracellular vesicles in glioma: Current status and future perspectives. Acta Oncol. 2019, 58, 353–362. [Google Scholar] [CrossRef]
- EV-TRACK Consortium; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Speicher, M. The biology of circulating tumor cells. Oncogene 2015, 35, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Bang-Christensen, S.R.; Pedersen, R.S.; Pereira, M.A.; Clausen, T.M.; Løppke, C.; Sand, N.T.; Ahrens, T.D.; Jørgensen, A.M.; Lim, Y.C.; Goksøyr, L.; et al. Capture and Detection of Circulating Glioma Cells Using the Recombinant VAR2CSA Malaria Protein. Cells 2019, 8, 998. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Chekhonin, V.P. Circulating tumor cells and their advances to promote cancer metastasis and relapse, with focus on glioblastoma multiforme. Exp. Mol. Pathol. 2018, 105, 166–174. [Google Scholar] [CrossRef]
- Li, M.X.; Ren, X.H.; Jiang, H.; Yang, K.Y.; Lin, S.; Cui, Y. Identification of circulating tumor cells in peripheral blood for gliomas by detection of aneuploid cells. Zhonghua Yi Xue Za Zhi 2019, 99, 1184–1188. [Google Scholar] [PubMed]
- Liu, T.; Xu, H.; Huang, M.; Ma, W.; Saxena, D.; Lustig, R.A.; Alonso-Basanta, M.; Zhang, Z.; Rourke, D.M.O.; Zhang, L.; et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Res. 2018, 78, 6632–6642. [Google Scholar] [CrossRef]
- Lombard, A.; Goffart, N.; Rogister, B. Glioblastoma Circulating Cells: Reality, Trap or Illusion? Stem Cells Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.-S.; Sheridan, M.; Van Gelder, J.; Darwish, B.; McKechnie, S.; et al. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- Müller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M.; et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014, 6, 247ra101. [Google Scholar] [CrossRef]
- Qi, Y.; Sun, Q.; Deng, G.; Zhang, H.; Xu, Y.; Li, Y.; Huang, S.; Li, Y.; Ye, Z.; Wang, Y.; et al. Identifying circulating glioma cells and their clusters as diagnostic markers by a novel detection platform. Clin. Transl. Med. 2021, 11, e318. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.P.; Nahed, B.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Rothenberg, S.M.; Sequist, L.V.; et al. Brain Tumor Cells in Circulation Are Enriched for Mesenchymal Gene Expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Zachariah, M.A.; Oliveira-Costa, J.P.; Carter, B.S.; Stott, S.L.; Nahed, B.V. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro-Oncology 2018, 20, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.; Pienta, K.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- Touat, M.; Duran-Peña, A.; Alentorn, A.; Lacroix, L.; Massard, C.; Idbaih, A. Emerging Circulating Biomarkers in Glioblas-toma: Promises and Challenges. Expert Rev. Mol. Diagn. 2015, 15, 1311–1323. [Google Scholar] [CrossRef] [PubMed]

| Families | Types |
|---|---|
| Adult-type diffuse gliomas | Astrocytoma, IDH-mutant |
| Oligodendroglioma, IDH-mutant, and 1p/19q-codeleted | |
| Glioblastoma, IDH-wildtype | |
| Pediatric-type diffuse low-grade gliomas | Diffuse astrocytoma, MYB- or MYBL1-altered |
| Angiocentric glioma | |
| Polymorphous low-grade neuroepithelial tumor of the young | |
| Diffuse low-grade glioma, MAPK pathway-altered | |
| Pediatric-type diffuse high-grade gliomas | Diffuse midline glioma, H3 K27-altered |
| Diffuse hemispheric glioma, H3 G34-mutant | |
| Diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype | |
| Infant-type hemispheric glioma | |
| Circumscribed astrocytic gliomas | Pilocytic astrocytoma |
| High-grade astrocytoma with piloid features | |
| Pleomorphic xanthoastrocytoma | |
| Subependymal giant cell astrocytoma | |
| Chordoid glioma | |
| Astroblastoma, MN1-altered |
| Biomarker | Prognostic Value | OS (HR) 95% CI | PFS (HR) 95% CI | n | CNS WHO Grade | Reference |
|---|---|---|---|---|---|---|
| IDH mutation | Positive | 0.241 (0.107–0.544) | NA | 98 | 4 | [40] |
| IDH mutation | Positive | 0.33 (0.25–0.42) | 0.38 (0.21–0.68) | 2190 | AGG | [38] |
| IDH mutation | Positive | 0.20 (0.06–0.58) | 0.14 (0.05–0.38) | 108 | LGG | [34] |
| 1p/19q codeletion | Positive | 0.33 | 0.34 | 79 | 4 | [33] |
| 1p/19q codeletion | Positive | 0.30 (0.12–0.75) | 0.34 (0.17–0.69) | 203 | 2 | [54] |
| 1p/19q codeletion | Positive | 0.43 (0.35–0.53) | 0.63 (0.52–0.76) | 3408 | AGG | [55] |
| MAPK pathway alterations | Positive | 0.19 (0.07–0.50) | NA | 64 | AGG | [56] |
| ATRX | Positive | 0.36 (0.17–0.81) | NA | 1206 | AGG | [53] |
| mMGMT (TMZ-treated PT) | Positive | 0.59 (0.37–0.94) | NA | 274 | 4 | [36] |
| mMGMT | Positive | 0.42 (0.38–0.45) | 0.43 (0.38–0.48) | 5103 | 4 | [57] |
| mMGMT (TMZ-treated PT) | Positive | 0.46 (0.41–0.52) | 0.48 (0.40–0.57) | 7888 | 4 | [58] |
| mMGMT (TMZ-free) | No effect | 0.97 (0.91–1.03) | 0.76 (0.57–1.02) | 7888 | 4 | [58] |
| TERT-mut (mMGMT PT) | Positive | 0.73 (0.55–0.98) | NA | 2819 | AGG | [59] |
| TERT-mut (MGMT-free) | Negative | 1.86 (1.54–2.26) | NA | 2819 | AGG | [59] |
| TERT-mut | Negative | 1.37 (1.08–1.76) | 1.37 (1.08–1.72) | 785 | AGG | [60] |
| TERT-mut | Negative | 1.38 (1.15–1.67) | 1.31 (1.06–1.63) | 11519 | LGG | [61] |
| High cfDNA | Negative | 1.82 (0.61–5.42) | NA | 42 | 4 | [62] |
| High ctDNA | Negative | 2.43 (1.19–4.95) | 2.19 (1.26–3.81) | 62 | 4 | [63] |
| High ctDNA | Negative | NA | NA | 85 | AGG | [64] |
| H3F3A | Negative | 4.27 (1.3–14.5) | NA | 42 | 4 | [45] |
| CDKN2A | Negative | 2.2 | 2.1 | 2193 | AGG | [65] |
| EGFR amplification | Negative | 0.43 (0.24–0.77) | NA | 718 | LGG | [66] |
| lncRNA | Negative | 2.09 (1.68–2.58) | NA | 1415 | AGG | [67] |
| Elevated serum YKL-40 | Negative | 1.4 (1.2–2.0) | NA | 343 | HGG | [68] |
| miRNA-221 upreg. | Negative | 1.66 (1.34–2.04) | 1.14 (1.02–1.26) | 1069 | AGG | [69] |
| miRNA-221 upreg. | Negative | 2.13 (1.05–4.31) | NA | 50 | 4 | [70] |
| miRNA-221 upreg. | Negative | 1.269 (1.054–1.527) | NA | 4708 | AGG | [71] |
| miRNA-155 upreg. | Negative | 1.4 (1.19–1.63) | NA | 1259 | AGG | [72] |
| miRNA-21 upreg. | Negative | 1.91 (1.34–2.73) | 1.23 (0.41–3.72) | 1059 | AGG | [73] |
| miRNA-21 upreg. | Negative | 1.681 (1.265–2.097) | NA | 1681 | AGG | [74] |
| miRNA-21 upreg. | Negative | 1.591 (1.278–1.981) | NA | 4708 | AGG | [71] |
| miRNA-222 upreg. | Negative | 2.09 (1.00–4.37) | NA | 50 | 4 | [70] |
| miRNA-222 upreg. | Negative | 1.72 (1.31–2.26) | 1.02 (0.86–1.22) | 1546 | AGG | [75] |
| miRNA-15b upreg. | Negative | 1.584 (1.199–2.092) | NA | 4708 | AGG | [71] |
| miRNA-148a upreg. | Negative | 1.122 (1.023–1.231) | NA | 4708 | AGG | [71] |
| miRNA-196 upreg. | Negative | 1.877 (1.033–3.411) | NA | 4708 | AGG | [71] |
| miRNA-210 upreg. | Negative | 1.251 (1.010–1.550) | NA | 4708 | AGG | [71] |
| miRNAs Upregulated in Gliomas | miRNAs Downregulated in Gliomas | ||||
|---|---|---|---|---|---|
| miRNA | Sample | References | miRNA | Sample | References |
| miRNA-21 | tissue, blood, CSF | [71,74,199,211,212,213,214,215,216,217,218,219] | miRNA-137 | tissue | [216,219,220] |
| miRNA-221 | tissue, blood | [71,213,214,221,222] | miRNA-342-3p | tissue, blood | [197,219] |
| miRNA-10 | tissue, blood, CSF | [204,216,220,223] | miRNA-124 | tissue | [71,217] |
| miRNA-222 | tissue, blood, CSF | [70,75,198] | miRNA-181a, miRNA-181b, and miRNA-181c | tissue | [221] |
| miRNA-210 | tissue, blood | [71,220] | miRNA-31 | tissue | [216] |
| miRNA-17 | tissue | [214] | miRNA-101 | tissue | [216] |
| miRNA-155 | tissue | [72,215] | miRNA-222 | tissue | [216] |
| miRNA-576-5p | blood | [224] | miRNA-330 | tissue | [216] |
| miRNA-340 | blood | [224] | miRNA-7 | tissue | [216] |
| miRNA-626 | blood | [224] | miRNA-7-5P | blood | [224] |
| miRNA-630 | tissue | [215] | let-7g-5p | blood | [224] |
| miRNA-1260 | tissue | [215] | miRNA-320 | blood | [224] |
| miRNA-542-5p | tissue | [215] | miRNA-125b | blood | [200] |
| miRNA-142-5p | tissue | [215] | miRNA-29 | blood | [220] |
| miRNA-106a-5p | blood | [220] | miRNA-197 | blood | [220] |
| miRNA-185 | blood | [220] | miRNA-205 | blood | [220] |
| miRNA-125 | CSF, blood | [198] | miRNA-485 | blood | [220] |
| miRNA-15b | tissue | [71] | miRNA-106 | tissue | [71] |
| miRNA-148a | tissue | [71] | |||
| miRNA-196 | tissue | [71] | |||
| miRNA-15a | tissue | [217] | |||
| miRNA-16 | tissue | [217] | |||
| miRNA-23a | tissue | [217] | |||
| miRNA-9 | tissue | [217] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. https://doi.org/10.3390/ijms221910373
Śledzińska P, Bebyn MG, Furtak J, Kowalewski J, Lewandowska MA. Prognostic and Predictive Biomarkers in Gliomas. International Journal of Molecular Sciences. 2021; 22(19):10373. https://doi.org/10.3390/ijms221910373
Chicago/Turabian StyleŚledzińska, Paulina, Marek G. Bebyn, Jacek Furtak, Janusz Kowalewski, and Marzena A. Lewandowska. 2021. "Prognostic and Predictive Biomarkers in Gliomas" International Journal of Molecular Sciences 22, no. 19: 10373. https://doi.org/10.3390/ijms221910373
APA StyleŚledzińska, P., Bebyn, M. G., Furtak, J., Kowalewski, J., & Lewandowska, M. A. (2021). Prognostic and Predictive Biomarkers in Gliomas. International Journal of Molecular Sciences, 22(19), 10373. https://doi.org/10.3390/ijms221910373






