FISH and Chimps: Insights into Frequency and Distribution of Sperm Aneuploidy in Chimpanzees (Pan troglodytes)
Abstract
:1. Introduction
2. Results
2.1. Chimpanzee Biological Data
2.2. DNA Probes
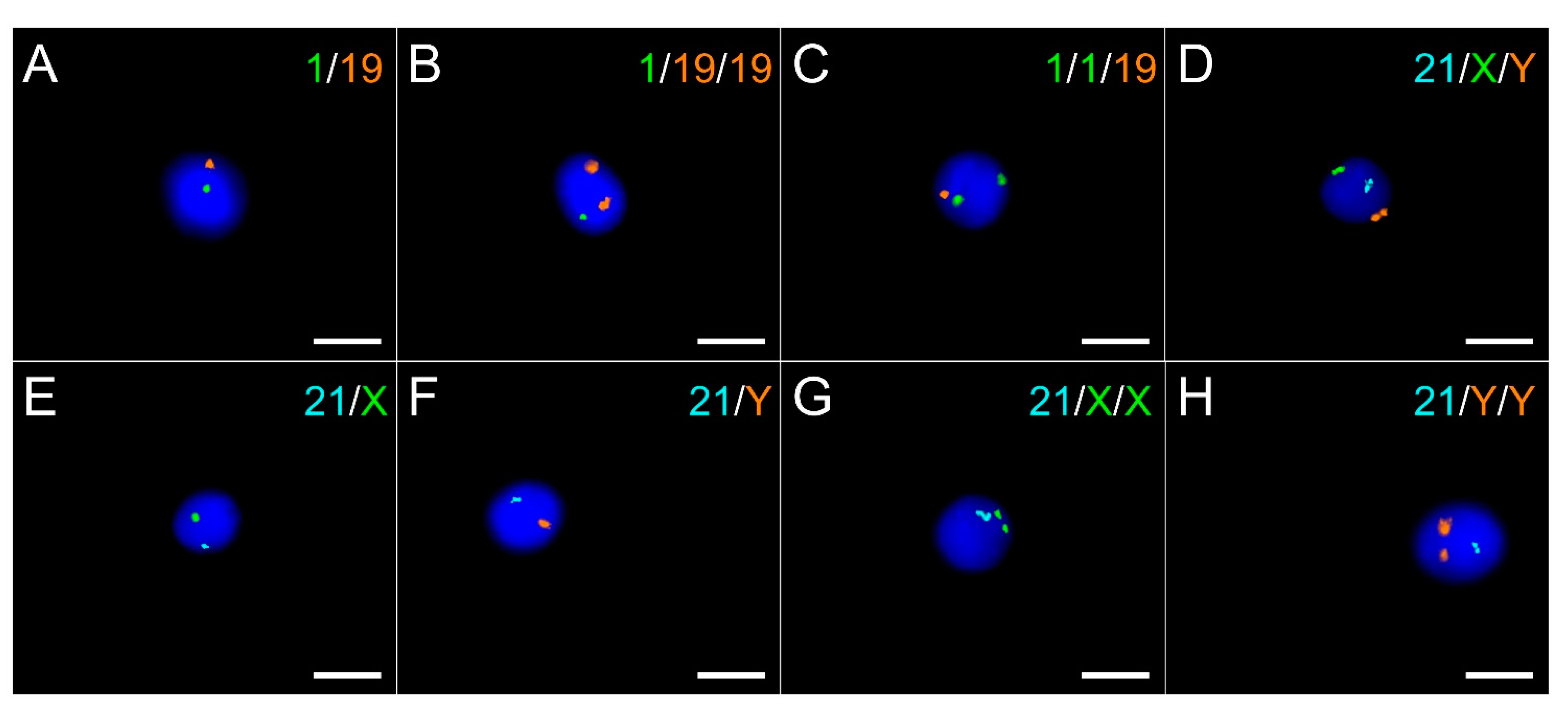
2.3. Sperm Aneuploidy
2.4. Interspecies Comparison of Sperm Aneuploidy Rates
3. Discussion
4. Materials and Methods
4.1. Animals and Sample Collection
4.2. Karyotype
4.3. Sperm Preparation
4.4. DNA Probes
4.5. FISH and Scoring
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ART | assisted reproduction techniques |
| BAC | bacterial artificial chromosome |
| df | degrees of freedom |
| DNA | deoxyribonucleic acid |
| EAZA | European Association of Zoos and Aquaria |
| FISH | fluorescence in situ hybridization |
| HSA | Homo sapiens |
| MMU | Macaca mulatta |
| NOR | nucleolus organizer regions |
| NS | not significant |
| PTR | Pan troglodytes |
| SA | spectrum aqua |
| SE | standard error |
| SG | spectrum green |
| SO | spectrum orange |
| WHO | World Health Organization |
References
- Fenster, C.; Ballou, J.; Dudash, M.; Eldridge, M.; Frankham, R.; Lacy, R.; Ralls, K.; Sunnucks, P. Conservation and Genetics. Yale J. Biol. Med. 2018, 91, 491–501. [Google Scholar]
- International Union for Conservation of Nature—IUCN Website. Available online: https://www.iucn.org/fr (accessed on 2 February 2021).
- Estrada, A.; Garber, P.; Rylands, A.; Roos, C.; Fernandez-Duque, E.; Di Fiore, A.; Nekaris, K.; Nijman, V.; Heymann, E.; Lambert, J.; et al. Impending extinction crisis of the world’s primates: Why primates matter. Sci. Adv. 2017, 3, e1600946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osada, N. Genetic diversity in humans and non-human primates and its evolutionary consequences. Genes Genet. Syst. 2015, 90, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSalle, R.; Amato, G. The expansion of conservation genetics. Nat. Rev. Genet. 2004, 5, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Arandjelovic, M.; Vigilant, L. Non-invasive genetic censusing and monitoring of primate populations. Am. J. Primatol. 2018, 80, e22743. [Google Scholar] [CrossRef]
- Andrabi, S.; Maxwell, W. A review on reproductive biotechnologies for conservation of endangered mammalian species. Anim. Reprod. Sci. 2007, 99, 223–243. [Google Scholar] [CrossRef]
- Comizzoli, P.; Holt, W. Breakthroughs and new horizons in reproductive biology of rare and endangered animal species. Biol. Reprod. 2019, 101, 514–525. [Google Scholar] [CrossRef] [Green Version]
- Martinez, G.; Garcia, C. Sexual selection and sperm diversity in primates. Mol. Cell. Endocrinol. 2020, 518, 110974. [Google Scholar] [CrossRef]
- Villagómez, D.; Pinton, A. Chromosomal abnormalities, meiotic behavior and fertility in domestic animals. Cytogenet. Genome Res. 2008, 120, 69–80. [Google Scholar] [CrossRef]
- Villagómez, D.; Parma, P.; Radi, O.; Di Meo, G.; Pinton, A.; Iannuzzi, L.; King, W. Classical and Molecular Cytogenetics of Disorders of Sex Development in Domestic Animals. Cytogenet. Genome Res. 2009, 126, 110–131. [Google Scholar] [CrossRef]
- Ramasamy, R.; Scovell, J.; Kovac, J.; Cook, P.; Lamb, D.; Lipshultz, L. Fluorescence in situ hybridization detects increased sperm aneuploidy in men with recurrent pregnancy loss. Fertil. Steril. 2015, 103, 906–909.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raudsepp, T.; Chowdhary, B. Chromosome Aberrations and Fertility Disorders in Domestic Animals. Annu. Rev. Anim. Biosci. 2016, 4, 15–43. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, L. Sperm genetic abnormalities and their contribution to embryo aneuploidy & miscarriage. Best Pract. Res. Clin. Endocrinol. Metab. 2020, 34, 101477. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Weick, R.; Knobil, E.; Wolman, S.; Gorstein, F. An X-O Anomaly and Ovarian Dysgenesis in a Rhesus Monkey. Folia Primatol. 1973, 19, 24–27. [Google Scholar] [CrossRef]
- Ruppenthal, G.; Caffery, S.; Goodlin, B.; Sackett, G.; Vigfusson, N.; Peterson, V. Pigtailed macaques (Macaca nemestrina) with trisomy X manifest physical and mental retardation. Am. J. Ment. Defic. 1983, 87, 471–476. [Google Scholar]
- Reyes, F.; Osborn, R.; Fuller, G.; Hobson, W.; Greenberg, C.; Ray, M.; Thliveris, J.; Faiman, C. Gonadal dysgenesis with X-monosomy in a cynomolgus monkey (Macaca fascicularis). Am. J. Primatol. 1990, 22, 51–59. [Google Scholar] [CrossRef]
- Moore, C.; Leland, M.; Brzyski, R.; McKeand, J.; Witte, S.; Rogers, J. A baboon (Papio hamadryas) with an isochromosome for the long arm of the X. Cytogenet. Genome Res. 1998, 82, 80–82. [Google Scholar] [CrossRef]
- Ruppenthal, G.; Moore, C.; Best, R.; Walker-Gelatt, C.; Delio, P.; Sackett, G. Trisomy 16 in a Pigtailed Macaque (M. nemestrina) With Multiple Anomalies and Developmental Delays. Am. J. Ment. Retard. 2004, 109, 9. [Google Scholar] [CrossRef]
- Dudley, C.; Hubbard, G.; Moore, C.; Dunn, B.; Raveendran, M.; Rogers, J.; Nathanielsz, P.; McCarrey, J.; Schlabritz-Loutsevitch, N. A male baboon (Papio hamadryas) with a mosaic 43,XXY/42,XY karyotype. Am. J. Med. Genet. Part A 2005, 140A, 94–97. [Google Scholar] [CrossRef]
- Hassanane, M.; Kovacs, A.; Laurent, P.; Lindblad, K.; Gustavsson, I. Simultaneous detection of X- and Y-bearing bull spermatozoa by double colour fluorescence in situ hybridization. Mol. Reprod. Dev. 1999, 53, 407–412. [Google Scholar] [CrossRef]
- Nicodemo, D.; Pauciullo, A.; Castello, A.; Roldan, E.; Gomendio, M.; Cosenza, G.; Peretti, V.; Perucatti, A.; Di Meo, G.; Ramunno, L.; et al. X-Y Sperm Aneuploidy in 2 Cattle (Bos taurus) Breeds as Determined by Dual Color Fluorescent in situ Hybridization (FISH). Cytogenet. Genome Res. 2009, 126, 217–225. [Google Scholar] [CrossRef]
- Pauciullo, A.; Cosenza, G.; Peretti, V.; Iannuzzi, A.; Di Meo, G.; Ramunno, L.; Iannuzzi, L.; Rubes, J.; Di Berardino, D. Incidence of X-Y aneuploidy in sperm of two indigenous cattle breeds by using dual color fluorescent in situ hybridization (FISH). Theriogenology 2011, 76, 328–333. [Google Scholar] [CrossRef]
- Di Berardino, D.; Vozdova, M.; Kubickova, S.; Cernohorska, H.; Coppola, G.; Coppola, G.; Enne, G.; Rubes, J. Sexing river buffalo (Bubalus bubalis L.), sheep (Ovis aries L.), goat (Capra hircus L.), and cattle spermatozoa by double color FISH using bovine (Bos taurus L.) X- and Y-painting probes. Mol. Reprod. Dev. 2003, 67, 108–115. [Google Scholar] [CrossRef]
- Bugno-Poniewierska, M.; Kozub, D.; Pawlina, K.; Tischner, M.; Tischner, M.; Słota, E.; Wnuk, M. Determination of the Correlation Between Stallion’s Age and Number of Sex Chromosome Aberrations in Spermatozoa. Reprod. Domest. Anim. 2011, 46, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Bugno, M.; Jablonska, Z.; Tischner, M.; Klukowska-Rötzler, J.; Pienkowska-Schelling, A.; Schelling, C.; Slota, E. Detection of Sex Chromosome Aneuploidy in Equine Spermatozoa Using Fluorescence in Situ Hybridization. Reprod. Domest. Anim. 2009, 45, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Rubeš, J.; Vozdová, M.; Kubíčková, S. Aneuploidy in pig sperm: Multicolor fluorescence in situ hybridization using probes for chromosomes 1, 10, and Y. Cytogenet. Genome Res. 1999, 85, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Orsztynowicz, M.; Pawlak, P.; Oleś, D.; Kubickova, S.; Lechniak, D. Low incidence of chromosome aberrations in spermatozoa of fertile boars. Reprod. Biol. 2011, 11, 224–235. [Google Scholar] [CrossRef]
- Komaki, H.; Oi, M.; Suzuki, H. Detection of sex chromosome aneuploidy in dog spermatozoa by triple color fluorescence in situ hybridization. Theriogenology 2014, 82, 652–656. [Google Scholar] [CrossRef]
- O’Brien, J.; Stojanov, T.; Heffernan, S.; Hollinshead, F.; Vogelnest, L.; Chis Maxwell, W.; Evans, G. Flow cytometric sorting of non-human primate sperm nuclei. Theriogenology 2005, 63, 246–259. [Google Scholar] [CrossRef]
- Froenicke, L.; Hung, P.; VandeVoort, C.; Lyons, L. Development of a non-human primate sperm aneuploidy assay tested in the rhesus macaque (Macaca mulatta). MHR Basic Sci. Reprod. Med. 2007, 13, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Morrell, J.; Hodges, J. Cryopreservation of non-human primate sperm: Priorities for future research. Anim. Reprod. Sci. 1998, 53, s0378–s4320. [Google Scholar] [CrossRef]
- Netten, H.; Young, I.; van Vliet, L.; Tanke, H.; Vroljik, H.; Sloos, W. FISH and chips: Automation of fluorescent dot counting in interphase cell nuclei. Cytometry 1997, 28, 1–10. [Google Scholar] [CrossRef]
- Carrell, D.; Emery, B. Use of automated imaging and analysis technology for the detection of aneuploidy in human sperm. Fertil. Steril. 2008, 90, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Molina, Ò.; Sarrate, Z.; Vidal, F.; Blanco, J. FISH on sperm: Spot-counting to stop counting? Not yet. Fertil. Steril. 2009, 92, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Gillois, P.; Le Mitouard, M.; Borye, R.; Esquerré-Lamare, C.; Satre, V.; Bujan, L.; Hennebicq, S. FISH and tips: A large scale analysis of automated versus manual scoring for sperm aneuploidy detection. Basic Clin. Androl. 2013, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammers, J.; Splingart, C.; Barrière, P.; Jean, M.; Fréour, T. Double-blind prospective study comparing two automated sperm analyzers versus manual semen assessment. J. Assist. Reprod. Genet. 2013, 31, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.; Barber, G.; Casper, J.; Clawson, H.; Diekhans, M.; Gonzalez, J.; Hinrichs, A.; Lee, B.; Nassar, L.; Powell, C.; et al. UCSC Genome Browser enters 20th year. Nucleic Acids Res. 2019, 48, 756–761. [Google Scholar] [CrossRef]
- Kasai, F.; Takahashi, E.; Koyama, K.; Terao, K.; Suto, Y.; Tokunaga, K.; Nakamura, Y.; Hirai, M. Comparative FISH mapping of the ancestral fusion point of human chromosome 2. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2000, 8, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, D.; Fortun, J.; Tempest, H. Meiotic nondisjunction and sperm aneuploidy in humans. Reproduction 2018, 157, 15–31. [Google Scholar] [CrossRef] [Green Version]
- Schmickel, R.; Gonzalez, I.; Erickson, J. Nucleolus Organizing Genes on Chromosome 21: Recombination and Nondisjunction. Ann. N. Y. Acad. Sci. 1985, 450, 121–131. [Google Scholar] [CrossRef]
- García, M.; Dietrich, A.; Pujol, R.; Egozcue, J. Nucleolar structures in chromosome and SC preparations from human oocytes at first meiotic prophase. Hum. Genet. 1989, 82, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Ford, J.; Lester, P. Factors affecting the displacement of human chromosomes from the metaphase plate. Cytogenet. Genome Res. 1982, 33, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, E.; Rademaker, A.; Martin, R. Aneuploidy in human sperm: The use of multicolor FISH to test various theories of nondisjunction. Am. J. Hum. Genet. 1996, 58, 356–362. [Google Scholar]
- Ferlin, A.; Garolla, A.; Foresta, C. Chromosome abnormalities in sperm of individuals with constitutional sex chromosomal abnormalities. Cytogenet. Genome Res. 2005, 111, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cruz, R.; Casanovas, A.; Brieno-Enriquez, M.; Robles, P.; Roig, I.; Pujol, A.; Cabero, L.; Durban, M.; Garcia Caldes, M. Cytogenetic analyses of human oocytes provide new data on non-disjunction mechanisms and the origin of trisomy 16. Hum. Reprod. 2009, 25, 179–191. [Google Scholar] [CrossRef] [Green Version]
- Harton, G.; Tempest, H. Chromosomal disorders and male infertility. Asian J. Androl. 2011, 14, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Uroz, L.; Templado, C. Meiotic non-disjunction mechanisms in human fertile males. Hum. Reprod. 2012, 27, 1518–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Gao, H.; Zhao, Y.; Ma, S. Aneuploidy and DNA fragmentation in morphologically abnormal sperm. Int. J. Androl. 2009, 33, e163–e179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coban, O.; Serdarogullari, M.; Onar Sekerci, Z.; Bilgin, E.; Serakinci, N. Evaluation of the impact of sperm morphology on embryo aneuploidy rates in a donor oocyte program. Syst. Biol. Reprod. Med. 2018, 64, 169–173. [Google Scholar] [CrossRef]
- Nagaoka, S.; Hassold, T.; Hunt, P. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brieño-Enríquez, M.; Cohen, P. Double trouble in human aneuploidy. Nat. Genet. 2015, 47, 696–698. [Google Scholar] [CrossRef]
- Stevison, L.; Woerner, A.; Kidd, J.; Kelley, J.; Veeramah, K.; McManus, K.; Bustamante, C.; Hammer, M.; Wall, J. The Time Scale of Recombination Rate Evolution in Great Apes. Mol. Biol. Evol. 2015, 33, 928–945. [Google Scholar] [CrossRef] [Green Version]
- Auton, A.; Fledel-Alon, A.; Pfeifer, S.; Venn, O.; Segurel, L.; Street, T.; Leffler, E.; Bowden, R.; Aneas, I.; Broxholme, J.; et al. A Fine-Scale Chimpanzee Genetic Map from Population Sequencing. Science 2012, 336, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Scally, A.; Dutheil, J.; Hillier, L.; Jordan, G.; Goodhead, I.; Herrero, J.; Hobolth, A.; Lappalainen, T.; Mailund, T.; Marques-Bonet, T.; et al. Insights into hominid evolution from the gorilla genome sequence. Nature 2012, 483, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Liu, H.; Zhang, H.; Wang, L.; Li, M.; Cai, F.; Wang, X.; Wang, L.; Zhang, R.; Yang, S.; et al. Paternal USP26 mutations raise Klinefelter syndrome risk in the offspring of mice and humans. EMBO J. 2021, 40, e106864. [Google Scholar] [CrossRef]
- García-Ferreyra, J.; Hilario, R.; Dueñas, J. High percentages of embryos with 21, 18 or 13 trisomy are related to advanced paternal age in donor egg cycles. JBRA Assist. Reprod. 2018, 22, 26–34. [Google Scholar] [CrossRef]
- Robbins, W.; Elashoff, D.; Xun, L.; Jia, J.; Li, N.; Wu, G.; Wei, F. Effect of lifestyle exposures on sperm aneuploidy. Cytogenet. Genome Res. 2005, 111, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Walschaerts, M.; Le Mitouard, M.; Borye, R.; Thomas, C.; Auger, J.; Berthaut, I.; Brugnon, F.; Daudin, M.; Moinard, N.; et al. Impact of Hodgkin or non-Hodgkin lymphoma and their treatments on sperm aneuploidy: A prospective study by the French CECOS network. Fertil. Steril. 2017, 107, 341–350.e5. [Google Scholar] [CrossRef] [Green Version]
- Losdat, S.; Chang, S.; Reid, J. Inbreeding depression in male gametic performance. J. Evol. Biol. 2014, 27, 992–1011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhai, L.; Fang, Z.; Li, A.; Qiu, Y.; Liu, Y. Impact of a mild scrotal heating on sperm chromosomal abnormality, acrosin activity and seminal alpha-glucosidase in human fertile males. Andrologia 2018, 50, e12985. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.; Young, H.; Grandjean, P.; Halling, J.; Petersen, M.; Martenies, S.; Karimi, P.; Weihe, P. Sperm Aneuploidy in Faroese Men with Lifetime Exposure to Dichlorodiphenyldichloroethylene (p,p’-DDE) and Polychlorinated Biphenyl (PCB) Pollutants. Environ. Health Perspect. 2016, 124, 951–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radwan, M.; Jurewicz, J.; Sobala, W.; Brzeźnicki, S.; Radwan, P.; Jakubowski, L.; Hawuła, W.; Ulańska, A.; Hanke, W. Human sperm aneuploidy after exposure to polycyclic aromatic hydrocarbons. Reprod. Fertil. Dev. 2016, 28, 1376. [Google Scholar] [CrossRef] [PubMed]
- Governini, L.; Guerranti, C.; De Leo, V.; Boschi, L.; Luddi, A.; Gori, M.; Orvieto, R.; Piomboni, P. Chromosomal aneuploidies and DNA fragmentation of human spermatozoa from patients exposed to perfluorinated compounds. Andrologia 2014, 47, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Jurewicz, J.; Radwan, M.; Wielgomas, B.; Klimowska, A.; Kałużny, P.; Radwan, P.; Jakubowski, L.; Hanke, W. Environmental exposure to parabens and sperm chromosome disomy. Int. J. Environ. Health Res. 2017, 27, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Perry, M. Effects of environmental and occupational pesticide exposure on human sperm: A systematic review. Hum. Reprod. Update 2008, 14, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Rademaker, A.; Spriggs, E.; Ko, E.; Martin, R. Reliability of estimates of diploid human spermatozoa using multicolour fluorescence in-situ hybridization. Hum. Reprod. 1997, 12, 77–79. [Google Scholar] [CrossRef] [Green Version]
- Rubes, J.; Vozdova, M.; Oracova, E.; Perreault, S. Individual variation in the frequency of sperm aneuploidy in humans. Cytogenet. Genome Res. 2005, 111, 229–236. [Google Scholar] [CrossRef]
- Tempest, H.; Ko, E.; Rademaker, A.; Chan, P.; Robaire, B.; Martin, R. Intra-individual and inter-individual variations in sperm aneuploidy frequencies in normal men. Fertil. Steril. 2009, 91, 185–192. [Google Scholar] [CrossRef]

| Individual 1 | Individual 2 | Literature Range or Mean (n = 69) † | ||
|---|---|---|---|---|
| Ejaculate | 1 | 2 | 1 | |
| Sperm parameters | ||||
| Sperm volume (µL) | 600 | 700 | 400 | 100–4400 |
| pH | 8 | 7.9 | - | - |
| Sperm concentration (106/mL) | 92 | 290 | 65 | 61–11300 |
| Vitality (% alive) | 62 | 77 | - | 72.13 |
| Sperm motility | ||||
| Total motility (%) | 55 | 72 | - | 3–93.2 |
| Average path velocity (µM/s) | 76.70 ± 40.67 | 107.32 ± 49.12 | - | 138 |
| Straight linear velocity (µM/s) | 58.15 ± 37.26 | 62.59 ± 48.62 | - | 34.3 |
| Curvilinear velocity (µM/s) | 128.43 ± 50.48 | 136.70 ± 53.25 | - | 110.3 |
| Amplitude of lateral head displacement (µM) | 7.32 ± 4.19 | 6.98 ± 4.57 | - | 5.2 |
| Sperm morphology | ||||
| Normal morphology (%) | 87 | 83 | 79 | 69.58 |
| Head defects (%) | 5 | 8 | 7 | - |
| Abnormal base (%) | 2 | 3 | 4 | - |
| Abnormal acrosome (%) | 2 | 4 | 2 | - |
| Macro head (%) | 0 | 1 | 0 | - |
| Thinned head (%) | 1 | 0 | 1 | - |
| Flagellum defects (%) | 8 | 9 | 14 | - |
| Coiled tail (%) | 4 | 5 | 6 | - |
| Destructured tail (%) | 2 | 1 | 1 | - |
| Multiple tail (%) | 1 | 0 | 0 | - |
| Simple bent tail (%) | 1 | 3 | 7 | - |
| Sperm morphometry | ||||
| Head length (µM ± SE) | 4.40 ± 0.22 | - | - | 4.8 |
| Head width (µM ± SE) | 2.79 ± 0.19 | - | - | 2.8 |
| Head area (µM ± SE) | 9.87 ± 0.80 | - | - | - |
| Head perimeter (µM ± SE) | 11.79 ± 0.44 | - | - | - |
| Head ellipticity (µM ± SE) | 1.58 ± 0.12 | - | - | 1.68 |
| Elongation (µM ± SE) | 0.22 ± 0.04 | - | - | 0.25 |
| Roughness (µM ± SE) | 0.89 ± 0.07 | - | - | - |
| Regularity (µM ± SE) | 0.98 ± 0.07 | - | - | - |
| Midpiece length (µM ± SE) | 5.67 ± 0.56 | - | - | 6.79 |
| Principal piece length (µM ± SE) | 54.03 ± 2.73 | - | - | - |
| Flagellum length (µM ± SE) | 59.71 ± 2.75 | - | - | 52.25 |
| Total length (µM ± SE) | 64.11 ± 2.75 | - | - | 59.30 |
| CHROMOSOME (PTR) | NUMBER OF DISOMIC CELLS (%) | NUMBER OF NUCLEI ANALYZED |
|---|---|---|
| 1 | 8 (0.080) | 10,027 |
| 2 | 5 (0.050) | 10,092 |
| 3 | 6 (0.060) | 10,013 |
| 4 | 6 (0.060) | 10,003 |
| 5 | 8 (0.079) | 10,121 |
| 6 | 6 (0.059) | 10,168 |
| 7 | 3 (0.030) | 10,059 |
| 8 | 2 (0.020) | 10,055 |
| 9 | 7 (0.069) | 10,092 |
| 10 | 5 (0.050) | 10,059 |
| 11 | 8 (0.078) | 10,278 |
| 12 | 5 (0.050) | 10,003 |
| 13 | 8 (0.079) | 10,168 |
| 14 | 5 (0.050) | 10,055 |
| 15 | 4 (0.040) | 10,121 |
| 16 | 9 (0.090) | 10,013 |
| 17 | 2 (0.020) | 10,003 |
| 18 | 7 (0.070) | 10,025 |
| 19 | 3 (0.030) | 10,025 |
| 20 | 7 (0.070) | 10,027 |
| 21 | 6 (0.060) | 10,003 |
| 22 | 8 (0.079) | 10,108 |
| 23 | 4 (0.039) | 10,298 |
| X/Y | 2 (0.198) | 10,108 |
| CHROMOSOME SET (PTR) | NUMBER OF DIPLOID CELLS (%) | NUMBER OF NUCLEI ANALYZED |
|---|---|---|
| 18/19 | 3 (0.030) | 10,025 |
| 7/10 | 5 (0.050) | 10,059 |
| 17/21 | 6 (0.060) | 10,003 |
| 6/13 | 10 (0.098) | 10,168 |
| 3/16 | 6 (0.060) | 10,013 |
| 11/23 | 8 (0.078) | 10,298 |
| 1/20 | 4 (0.040) | 10,027 |
| 2/9 | 6 (0.059) | 10,092 |
| 5/15 | 5 (0.049) | 10,121 |
| 4/12 | 4 (0.040) | 10,003 |
| 8/14 | 3 (0.030) | 10,055 |
| 22/X/Y | 4 (0.040) | 10,108 |
| MEAN ± SD | 64 (0.053) | 120,972 |
| HSA | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | XY | |
| disomy (%) | 0.088 | 0.089 | 0.200 | 0.078 | - | 0.040 | 0.050 | 0.038 | 0.094 | - | - | 0.145 | 0.109 | - | 0.346 | 0.209 | 0.170 | 0.077 | 0.430 | 0.120 | 0.154 | 0.370 | 0.267 | |
| PTR | 1 | 12 | 13 | 2 | 3 | 4 | 5 | 6 | 7 | 11 | 8 | 9 | 10 | 14 | 15 | 16 | 18 | 19 | 17 | 20 | 21 | 22 | 23 | XY |
| disomy (%) | 0.080 | 0.050 | 0.079 | 0.050 | 0.060 | 0.060 | 0.079 | 0.059 | 0.030 | 0.078 | 0.020 | 0.069 | 0.050 | 0.050 | 0.040 | 0.090 | 0.070 | 0.030 | 0.020 | 0.060 | 0.060 | 0.079 | 0.038 | 0.197 |
| MMU | 1 | 12 | 13 | 2 | 5 | 6 | 4 | 3q | 8 | 15 | 9 | 14 | 11 | 17 | 7q | 7p | 20 | 16 | 18 | 19 | 10q | 3p | 10p | XY |
| disomy (%) | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.040 | - | - | 0.060 | 0.040 | 0.040 | 0.030 | 0.140 | - | - | 0.190 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guyot, C.; Gandula, M.; Noordermeer, W.; François-Brazier, C.; Moigno, R.; Bessonnat, J.; Brouillet, S.; Dhellemmes, M.; Bidart, M.; Arnoult, C.; et al. FISH and Chimps: Insights into Frequency and Distribution of Sperm Aneuploidy in Chimpanzees (Pan troglodytes). Int. J. Mol. Sci. 2021, 22, 10383. https://doi.org/10.3390/ijms221910383
Guyot C, Gandula M, Noordermeer W, François-Brazier C, Moigno R, Bessonnat J, Brouillet S, Dhellemmes M, Bidart M, Arnoult C, et al. FISH and Chimps: Insights into Frequency and Distribution of Sperm Aneuploidy in Chimpanzees (Pan troglodytes). International Journal of Molecular Sciences. 2021; 22(19):10383. https://doi.org/10.3390/ijms221910383
Chicago/Turabian StyleGuyot, Charlotte, Marlène Gandula, Wendy Noordermeer, Céline François-Brazier, Rosemary Moigno, Julien Bessonnat, Sophie Brouillet, Magali Dhellemmes, Marie Bidart, Christophe Arnoult, and et al. 2021. "FISH and Chimps: Insights into Frequency and Distribution of Sperm Aneuploidy in Chimpanzees (Pan troglodytes)" International Journal of Molecular Sciences 22, no. 19: 10383. https://doi.org/10.3390/ijms221910383
APA StyleGuyot, C., Gandula, M., Noordermeer, W., François-Brazier, C., Moigno, R., Bessonnat, J., Brouillet, S., Dhellemmes, M., Bidart, M., Arnoult, C., Satre, V., Coutton, C., & Martinez, G. (2021). FISH and Chimps: Insights into Frequency and Distribution of Sperm Aneuploidy in Chimpanzees (Pan troglodytes). International Journal of Molecular Sciences, 22(19), 10383. https://doi.org/10.3390/ijms221910383






