Abstract
Preclinical studies have indicated that T-cell immunoglobulin and ITIM domain (TIGIT) can substantially attenuate anti-tumoral immune responses. Although multiple clinical studies have evaluated the significance of TIGIT in patients with solid cancers, their results remain inconclusive. Thus, we conducted the current systematic review and meta-analysis based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) to determine its significance in patients with solid cancers. We systematically searched the Web of Science, Embase, PubMed, and Scopus databases to obtain peer-reviewed studies published before September 20, 2020. Our results have shown that increased TIGIT expression has been significantly associated with inferior overall survival (OS) (HR = 1.42, 95% CI: 1.11–1.82, and p-value = 0.01). Besides, the level of tumor-infiltrating TIGIT+CD8+ T-cells have been remarkably associated inferior OS and relapse-free survival (RFS) of affected patients (HR = 2.17, 95% CI: 1.43–3.29, and p-value < 0.001, and HR = 1.89, 95% CI: 1.36–2.63, and p-value < 0.001, respectively). Also, there is a strong positive association between TIGIT expression with programmed cell death-1 (PD-1) expression in these patients (OR = 1.71, 95% CI: 1.10–2.68, and p-value = 0.02). In summary, increased TIGIT expression and increased infiltration of TIGIT+CD8+ T-cells can substantially worsen the prognosis of patients with solid cancers. Besides, concerning the observed strong association between TIGIT and PD-1, ongoing clinical trials, and promising preclinical results, PD-1/TIGIT dual blockade can potentially help overcome the immune-resistance state seen following monotherapy with a single immune checkpoint inhibitor in patients with solid cancers.
1. Introduction
Solid cancers have remained one of the daunting public health burdens worldwide [1]. Recent advances in immunotherapy have paved the way for introducing novel treatments for patients with solid tumors. However, the undesirable response rates of immunotherapeutic approaches have been a major obstacle for their translation into clinical practice for patients with solid cancers [2]. Thus, a better understanding of the cross-talk between immune cells and tumoral cells might provide valuable insights to ameliorate the response rates of affected patients to immunotherapeutic approaches.
The immunosuppressive tumor microenvironments of solid cancers might be critical in inhibiting the stimulation of effector immune cells [3,4]. Indeed, the immunosuppressive tumor microenvironment can exhaust effector immune cells and prevent tumor rejection [5]. Despite the ever-increasing FDA-approved immune checkpoint inhibitors for cancer patients, they have not completely restored the anti-tumoral immune responses in all solid cancers [6]. Therefore, there is a need to identify novel immune checkpoints in patients with solid cancers to restore the anti-tumoral immune responses.
The PD-1/programmed death-ligand 1 (PD-L1) inhibitory axis has been one of the well-studied inhibitory immune checkpoint axes in cancers; thus, targeting this axis via monoclonal antibodies was among the attempts to stimulate anti-tumoral immune responses [7]. This axis can be established between immune and tumor cells and can substantially contribute to immunosuppressive tumor microenvironment development [8]. Although monoclonal antibodies targeting this inhibitory axis have been promising for some solid cancers, like triple-negative breast cancer, they have not yielded meaningful results in other solid cancers, like glioblastoma [9,10]. The low response rate of some patients to anti-PD-1 might be stemmed from the fact that other inhibitory immune checkpoint molecules can also regulate anti-tumoral immune responses. Deng et al. have shown that CD8+ T-cell subpopulations widely express PD-1, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), T-cell immunoglobulin mucin-3 (TIM-3), and lymphocyte activation gene-3 (LAG-3), and TIGIT [11]. Kim et al. have indicated that the majority of exhausted CD8+ T-cells highly express CTLA-4, LAG-3, TIGIT, and TIM-3 [12]. Therefore, other inhibitory immune checkpoint molecules might contribute to maintaining the immunosuppressive tumor microenvironment following monotherapy with monoclonal antibodies targeting one inhibitory axis.
Firstly, Yu et al. have identified TIGIT as an inhibitory signal that can repress T-cell activation [13]. TIGIT can be overexpressed in CD8+ T-cells, regulatory T-cells (Tregs), CD4+ T-cells, and natural killer (NK) cells [6]. Zhang et al. have shown that the TIGIT blockade can enhance NK cell-mediated anti-tumoral immune responses and improves the response rates of monoclonal antibodies targeting the PD-1/PD-L1 axis [14]. Besides, Wu et al. have demonstrated that TIGIT can be overexpressed in tumor-infiltrating CD8+ and CD4+ T-cells, and there is a remarkable relationship between TIGIT with PD-1, LAG-3, and TIM-3 in the tumor-infiltrating CD4+ and CD8+ T-cells in animal models of head and neck squamous cell carcinomas [15]. Indeed, the co-expression of TIGIT with other inhibitory immune checkpoint inhibitors, e.g., PD-1 and V-domain immunoglobulin suppressor of T cell activation (VISTA), might provide ample opportunities to reverse the immune-resistance state implicated in the unfavorable response rate of immune therapy with one immune checkpoint inhibitor [16,17].
Herein, the current meta-analysis aims to systematically investigate the significance of TIGIT in patients with solid cancers. Besides offering an unbiased insight into the significance of TIGIT in patients with solid cancers, bridging the results of this study with the recent preclinical results that are discussed in the discussion section might provide ample opportunities to improve the response rate of solid cancer patients to immune checkpoint inhibitor-based therapies.
2. Material and Methods
This study was conducted according to the PRISMA statements [18].
2.1. The Strategy of the Systematic Search
The Web of Science, Embase, PubMed, and Scopus databases were systematically searched to obtain the peer-reviewed records published before 20 September 2020. For this purpose, the aforementioned databases were systematically searched with the following keywords: (“tumor” OR “tumour” OR “malignancy” OR “neoplasm” OR “neoplasia” OR “malignant” OR “carcinoma” OR “cancerous” OR “tumoral” OR “tumoural” OR “neoplastic”) and (“T-cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domain” OR “T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif (ITIM) domain” OR “T Cell Immunoreceptor With Ig and ITIM Domains” OR “T cell immunoglobulin and ITIM domain” OR “T-Cell Immunoreceptor With Ig and ITIM Domains” OR “T-cell immunoglobulin and ITIM domain” OR “V-Set and Transmembrane Domain-Containing Protein 3” OR “V-Set and Transmembrane Domain Containing 3” OR “V-Set and Immunoglobulin Domain-Containing Protein 9” OR “V-Set and Immunoglobulin Domain Containing 9” OR “VSIG9” OR “VSTM3” OR “WUCAM” OR “TIGIT”).
2.2. Study Selection and Data Extraction
Following the systematic search, the obtained records were reviewed in two phases. In phase I, the records were screened based on their titles/abstracts. In phase II, the full text of papers and their supplementary data were reviewed for consideration to be involved in the current study. Any disagreements were resolved via consulting with B.B and consensus.
2.3. Eligibility Criteria
Papers with the following eligibility criteria were included in the current study: (1) clinical studies, (2) studies with the objective of assessing TIGIT immune checkpoint in patients with solid cancers, (3) studies that evaluated the prognostic value of TIGIT immune checkpoint or the clinicopathological significance of TIGIT in patients with solid cancers, and (4) studies that were published in English.
2.4. Data Extraction
The following data were extracted from the included studies: (1) the first author, (2) the publication year, (3) the country, (4) the sample size, (5) male to female ratio, (6) median age, (7) high tumor stage/low tumor stage ratio, (8) the cancer therapy records of the patients, (9) TIGIT evaluation method, (10) TIGIT antibody ID, (11) the prognostic value of TIGIT immune checkpoint, (12) the clinicopathological significance of TIGIT, (13) the association between TIGIT and PD-1 immune checkpoints, and (14) the prognostic value of TIGIT+CD8+ tumor-infiltrating lymphocytes.
2.5. Assessing the Potential Risk of Bias among the Included Studies
To improve the transparency of the obtained results, we applied the Hayden et al. guideline to assess the quality of included prognostic studies [19]. We also applied the JBI critical appraisal checklist for evaluating the quality of studies concerning the clinicopathological significance of TIGIT [20].
2.6. Statistical Analysis
The analyses were performed by STATA16 (StataCorp, College Station, TX, USA). Random effect meta-analyses were conducted utilizing the REML [21]. The random-effect model was applied because there may be other unknown, unregistered/unpublished studies that we could not have access. The I-squared, H-squared, Tau-squared, and Cochran Q test statistics were performed to assess included studies’ heterogenicity. Regarding the I-squared, the value above 50% was considered as high heterogeneity, and the H-Squared = 1 was considered as homogeneity among included studies [22]. The funnel plots were provided to assess the asymmetry and publication bias. For the evaluation of bias, Egger’s and Begg’s tests were performed [23,24].
3. Results
3.1. Systematic Search
Our systematic search retrieved 1410 records. After removing duplicated studies, 884 records remained. Based on screening the title/abstract of the records, 712 papers did not meet the aforementioned inclusion criteria. In phase II, the full text of the remaining 172 studies and their supplementary data were reviewed. Finally, six studies met the abovementioned criteria and were included in the quantitative synthesis. The flowchart of literature inclusion and exclusion is shown in Figure 1.
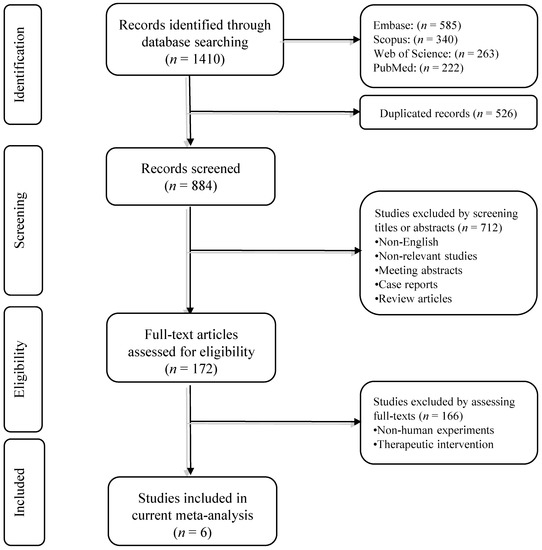
Figure 1.
The flowchart of the study selection process.
3.2. The Characteristic of Included Studies
The characteristic of the included studies is demonstrated in Table 1. The six clinical studies were published between 2018 and 2020. The studied solid cancers were esophageal squamous cell carcinoma [25], renal cell carcinoma [26], gastric adenocarcinoma [27], cutaneous melanoma [28], lung adenocarcinoma [29], and muscle-invasive bladder cancer [30]. Two studies only evaluated the clinicopathological significance of TIGIT in affected patients [26,27]. Except for the study by Lee et al., other included studied have ethical approvals [25,26,27,29,30]. The characteristics of the included studies are demonstrated in Table 1.

Table 1.
The characteristics of included studies.
3.3. The Clinicopathological Significance of TIGIT
Our results have shown no statistically significant associations between TIGIT expression with tumor size and tumor differentiation (HR = 2.84, 95% CI: 0.61–13.29, p-value = 0.18, and HR = 0.89, 95% CI: 0.36–2.24, p-value = 0.81, respectively) (Figure 2).
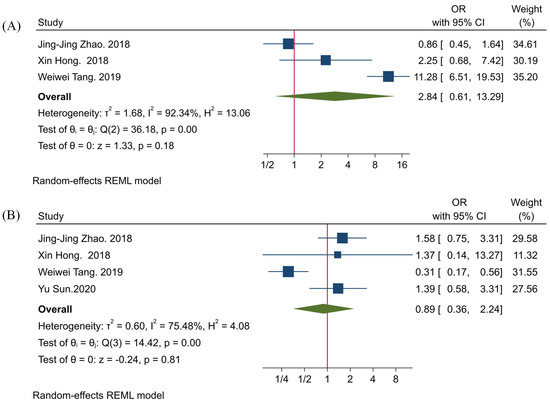
Figure 2.
Forest plots for evaluating the association between TIGIT expression and clinicopathological characteristics of affected patients. (A) tumor size, and (B) tumor differentiation.
3.4. The Association between TIGIT and PD-1
Our study has demonstrated that there is a significant association between the expression of TIGIT and PD-1 (OR = 1.71, 95% CI: 1.10–2.68, and p-value = 0.02) (Figure 3). Besides, our results have indicated that there is no significant heterogeneity among the included studies (p-value = 0.91, I2 = 0.00%, and H2 = 1.00) (Figure 3).
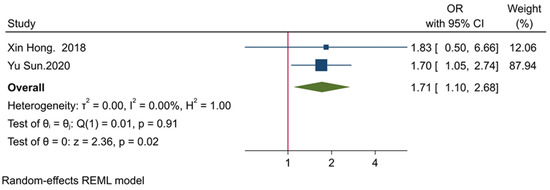
Figure 3.
Forest plot for evaluating the association between the expression of TIGIT and PD-1.
3.5. The Prognostic Value of TIGIT
Our results have shown that increased TIGIT expression is significantly associated with the inferior OS of affected patients (HR = 1.42, 95% CI: 1.11–1.82, and p-value = 0.01) (Figure 4). Also, our results have indicated that there is no significant heterogeneity among the included studies (p-value = 0.49, I2 = 2.24%, and H2 = 1.02) (Figure 4).
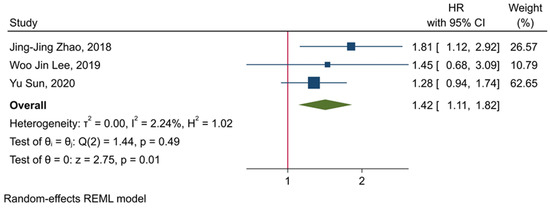
Figure 4.
Forest plot for evaluating the prognostic value of TIGIT expression in determining the OS of affected patients.
3.6. The Prognostic Value of Tumor-Infiltrating TIGIT+CD8+ T-Cells
Our results have shown that the level of tumor-infiltrating TIGIT+CD8+ T-cells is significantly associated the inferior OS and RFS of affected patients (HR = 2.17, 95% CI: 1.43–3.29, and p-value < 0.001, and HR = 1.89, 95% CI: 1.3–2.63, and p-value < 0.001, respectively) (Figure 5). Besides, our results have indicated that there is no significant heterogeneity among the included studies (p-value = 0.52, I2 = 0.00%, and H2 = 1.00 in the case of OS, and p-value = 0.56, I2 = 0.00%, and H2 = 1.00 in the case of RFS) (Figure 5).
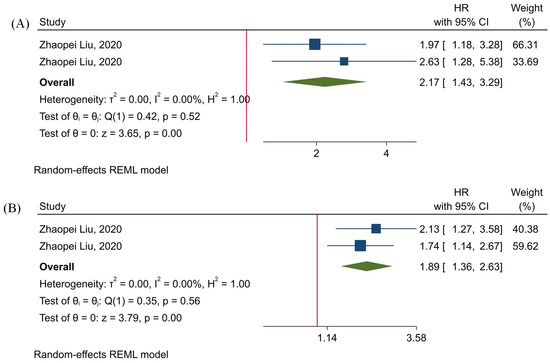
Figure 5.
Forest plots for assessing the prognostic value of tumor-infiltrating TIGIT+CD8+ T-cells. (A) OS, and (B) RFS.
3.7. Assessing Potential Bias among the Included Studies
The summaries of the evaluated quality of the included studies are demonstrated in Table 2 and Table 3. The main risk area has been about addressing the cofounders, which is stemmed from the nature of non-randomized studies. However, the quality of the included studies in other areas has been acceptable (Table 2 and Table 3). Overall, the quality of the included studies is acceptable.

Table 2.
Evaluating the potential bias in the included prognostic studies based on the Hayden et al. statements.

Table 3.
Evaluating the potential bias in the included studies based on the JBI critical appraisal checklist.
3.8. Evaluating Publication Bias
The Egger’s and Begg’s tests have demonstrated no significant publication bias (Figure 6).
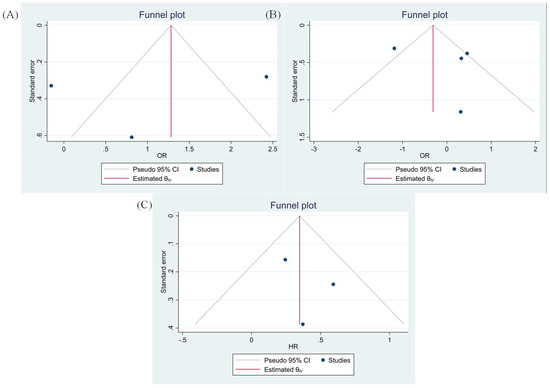
Figure 6.
Funnel plots based on TIGIT expression (A) tumor size (Egger test p-value = 0.769 and Begg’s test p-value = 0.149), (B) tumor differentiation (Egger test p-value = 0.579 and Begg’s test p-value = 0.367) and (C) OS (Egger test p-value = 0.500 and Begg’s test p-value = 0.500).
4. Discussion
Although multiple studies have investigated the prognostic value and clinicopathological significance of TIGIT in patients with solid cancers, their results remain inconclusive [25,26,27,28,29,30]. The current study is the first meta-analysis to study the significance of TIGIT in patients with solid cancers.
As a novel inhibitory immune checkpoint, TIGIT competes with co-stimulatory CD226 to bind with CD155 and CD112. TIGIT has a central role in attenuating immune responses. TIGIT can substantially impair dendritic cells via upregulating IL-10 expression [13]. IL-10 has been associated with decreased function of dendritic cells in developing anti-tumoral immune responses [31]. Besides, TIGIT can substantially inhibit NK cell-mediated anti-tumoral immune responses. Stanietsky et al. have shown that TIGIT can inhibit NK cells cytotoxicity, and its blockade can upregulate interferon-γ (IFN-γ) [32]. Meng et al. have reported that TIGIT+NK cells express fewer IFN-γ and tumor necrosis factor-α (TNF-α) than TIGIT-NK cells, and the high expression of TIGIT is associated with decreased function of NK cells and immune evasion of tumoral cells [33]. Moreover, TIGIT has been implicated in impairing the anti-tumoral immune responses of CD8+ T-cells. Weiling et al. have shown that TIGIT+ CD8 T-cells are functionally exhausted, and their proliferation is limited. Besides, tumoral cells can upregulate CD155 and facilitate CD8+ T-cells inactivation via the TIGIT/CD155 inhibitory axis. Also, silencing tumoral CD155 has been associated with increased expression of IFN-γ and restored metabolism of T-cells [34]. Furthermore, Zhou et al. have shown that tumor-intrinsic TIGIT can substantially inhibit NK-cells and CD8+ T-cells-mediated anti-tumoral immune responses and pave the way for tumor growth in vivo [35]. Consistent with these preclinical findings, our results have demonstrated that high expression of TIGIT is significantly associated with inferior OS in patients with solid cancers (HR = 1.42, 95% CI: 1.11–1.82, and p-value = 0.01). Besides, our results have indicated that increased infiltration of tumor-infiltrating TIGIT+CD8+ T-cells is remarkably associated inferior OS and RFS (HR = 2.17, 95% CI: 1.43–3.29, and p-value < 0.001, and HR = 1.89, 95% CI: 1.36–2.63, and p-value < 0.001, respectively). Therefore, TIGIT is a pivotal inhibitory immune checkpoint that its high expression in tumoral cells and cells residing in the tumor microenvironment can remarkably attenuate anti-tumoral immune responses.
Our results have indicated that the high expression of TIGIT might not be statistically associated with poor tumor differentiation and increased tumor size (both p-values > 0.05). However, the current meta-analysis has shown a significant association between TIGIT and PD-1 in patients with solid cancers (OR = 1.71, 95% CI: 1.10–2.68, and p-value = 0.02). Wu et al. have reported that anti-TIGIT monoclonal antibodies can substantially decrease arginase-1 transcription levels in myeloid-derived suppressor cells (MDSCs) [15]. Liu et al. have reported that anti-PD-1 monoclonal antibodies and PD-1 silencing of MDSCs can also substantially decrease arginase-1 expression in MDSCs [36]. Arginase-1 is an inhibitory factor that depletes L-arginine from the tumor microenvironment and induces T cell anergy [37]. Dufait et al. have shown that arginase-1 expression is elevated in tumor and in vitro generated MDSCs, and its inhibition can substantially increase the proliferation of T cells and decrease tumor volume in animal models [38]. MDSCs can remarkably express arginase-1 and contribute to the development of the immunosuppressive tumor microenvironment. Heuvers et al. have shown a remarkable correlation between arginase-1 and peripheral blood MDSCs [39]. Consistent with this, Ren et al. have indicated that early-stage MDSCs of peripheral blood and tumor-infiltrating early-stage MDSCs can upregulate arginase-1 [40]. Besides, the co-culture of MDSCs with tumoral cells can upregulate arginase-1 expression in MDSCs [41]. Although the exact mechanism of TIGIT and PD-1 co-expression has not been exactly identified in tumors, the currently available evidence indicates that TIGIT and PD-1 can both increase arginase-1 activity, which can ultimately lead to T cells anergy and the development of immunosuppressive tumor microenvironment. However, further studies are needed to elucidate the underlying mechanisms for the co-expression of TIGIT and PD-1 in cancer.
In line with our obtained results, Hung et al. have shown that TIGIT and PD-1 co-expression are substantially upregulated in tumor-infiltrating CD8+ T-cells in glioma animal models [16]. In patients with bladder cancer, TIGIT expression has also been predominantly co-expressed on PD-1+ tumor-infiltrating CD8+ T-cells. Moreover, TIGIT blockade can improve the efficacy of anti-PD-1 therapy and promote the stimulation of tumor-infiltrating CD8+ T-cells in patients with bladder cancer [42]. Besides, TIGIT/PD-1 dual blockade has remarkably improved anti-tumoral immune responses and promoted tumor rejection in lymphoma animal models [10]. Furthermore, the dual blockade of the immune checkpoint axes of PD-1 and TIGIT has been superior in rejecting melanoma and non-small-cell lung carcinoma [43,44]. Although our results have only shown a strong positive association between PD-1 and TIGIT expression, the results of the aforementioned studies indicate that dual PD-1/TIGIT blockade is superior in tumor rejection, which might imply the strong positive association between PD-1 and TIGIT in the tumor microenvironment of solid cancers. Also, multiple ongoing clinical trials investigate the efficacy of dual TIGIT/PD-1 blockade in various solid cancers (Table 4).

Table 4.
The current clinical trials for targeting TIGIT immune checkpoint.
The current study has some strengths. First, despite the various types of studied solid cancers in the included studies, our presented results have been homogenous, and there has been no significant heterogeneity that can jeopardize the obtained results regarding TIGIT significance. Second, consistent with preclinical findings, the current meta-analysis has highlighted the remarkable association between TIGIT and PD-1 on the clinical scale. Third, concerning the fact that TIGIT can be overexpressed in various immune cells, the current study has demonstrated the prognostic value of tumor-infiltrating TIGIT+CD8+ T-cells in patients with solid cancers. Fourth, our study has bridged the preclinical findings with the clinical findings. However, this study has some limitations, as well. Only papers published in English have been included in the current study. The second limitation of our study stems from the very nature of non-randomized clinical trial studies, in which all the confounder variables might not be addressed.
5. Conclusions
The current study has shown that the elevated expression of TIGIT in the tumor microenvironment is associated with inferior OS. Besides, the increased infiltration of tumor-infiltrating TIGIT+CD8+ is associated with the worsened OS and RFS of affected patients. Concerning the observed results regarding the strong positive association between TIGIT and PD-1 and the promising results of preclinical studies, dual PD-1/TIGIT blockade can substantially help overcome the immune-resistance state in solid cancers. Consistent with the current evidence and the current trend in the clinical trials for solid cancer patients, dual PD-1/TIGIT blockade can potentially ameliorate the response rate of solid cancer patients to immune checkpoint inhibitors.
Author Contributions
N.H. contributed to the study selection; M.A.S. developed the systematic search, interpreted the results, and wrote most of the manuscript; M.A.J. and S.M.M. have conducted the analysis; N.K.A., O.B., R.F., Z.A., P.L. and N.A. have extracted the data from the included studies; N.S. and B.B. have supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We appreciate the researchers of the Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran. We also appreciate the researchers of the Research Center for Evidence-Based Medicine, Tabriz University of Medical Sciences, Tabriz, Iran (n = 67,322).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sambi, M.; Bagheri, L.; Szewczuk, M.R. Current challenges in cancer immunotherapy: Multimodal approaches to improve efficacy and patient response rates. J. Oncol. 2019, 2019, 4508794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, J.; Wickman, E.; DeRenzo, C.; Gottschalk, S. CAR T-cell Therapy for Solid Tumors: Bright Future or Dark Reality? Mol. Ther. 2020, 28, 2320–2339. [Google Scholar] [CrossRef] [PubMed]
- Lotfinejad, P.; Asghari Jafarabadi, M.; Abdoli Shadbad, M.; Kazemi, T.; Pashazadeh, F.; Sandoghchian Shotorbani, S.; Jadidi Niaragh, F.; Baghbanzadeh, A.; Vahed, N.; Silvestris, N. Prognostic role and clinical significance of tumor-infiltrating lymphocyte (TIL) and programmed death ligand 1 (PD-L1) expression in triple-negative breast cancer (TNBC): A systematic review and meta-analysis study. Diagnostics 2020, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, A.; Rostami, Z.; Safarpour, H.; Shadbad, M.A.; Nourbakhsh, N.S.; Argentiero, A.; Taefehshokr, S.; Tabrizi, N.J.; Kooshkaki, O.; Astamal, R.V. From Oncogenic Signaling Pathways to Single-Cell Sequencing of Immune Cells: Changing the Landscape of Cancer Immunotherapy. Molecules 2021, 26, 2278. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, N.; Derakhshani, A.; Kooshkaki, O.; Abdoli Shadbad, M.; Hajiasgharzadeh, K.; Baghbanzadeh, A.; Safarpour, H.; Mokhtarzadeh, A.; Brunetti, O.; Yue, S.C. Immune Checkpoints and CAR-T Cells: The Pioneers in Future Cancer Therapies? Int. J. Mol. Sci. 2020, 21, 8305. [Google Scholar] [CrossRef]
- Bardhan, K.; Anagnostou, T.; Boussiotis, V.A. The PD1: PD-L1/2 pathway from discovery to clinical implementation. Front. Immunol. 2016, 7, 550. [Google Scholar] [CrossRef] [Green Version]
- Shadbad, M.A.; Asadzadeh, Z.; Derakhshani, A.; Hosseinkhani, N.; Mokhtarzadeh, A.; Baghbanzadeh, A.; Hajiasgharzadeh, K.; Brunetti, O.; Argentiero, A.; Racanelli, V.; et al. A scoping review on the potentiality of PD-L1-inhibiting microRNAs in treating colorectal cancer: Toward single-cell sequencing-guided biocompatible-based delivery. Biomed. Pharmacother. 2021, 143, 112213. [Google Scholar] [CrossRef] [PubMed]
- Shadbad, M.A.; Asadzadeh, Z.; Hosseinkhani, N.; Derakhshani, A.; Alizadeh, N.; Brunetti, O.; Silvestris, N.; Baradaran, B. A Systematic Review of the Tumor-Infiltrating CD8+ T-Cells/PD-L1 Axis in High-Grade Glial Tumors: Toward Personalized Immuno-Oncology. Front. Immunol. 2021, 12, 634181. [Google Scholar] [CrossRef]
- Shadbad, M.A.; Safaei, S.; Brunetti, O.; Derakhshani, A.; Lotfinejad, P.; Mokhtarzadeh, A.; Hemmat, N.; Racanelli, V.; Solimando, A.G.; Argentiero, A. A Systematic Review on the Therapeutic Potentiality of PD-L1-Inhibiting MicroRNAs for Triple-Negative Breast Cancer: Toward Single-Cell Sequencing-Guided Biomimetic Delivery. Genes 2021, 12, 1206. [Google Scholar] [CrossRef]
- Deng, W.; Ma, Y.; Su, Z.; Liu, Y.; Liang, P.; Huang, C.; Liu, X.; Shao, J.; Zhang, Y.; Zhang, K.; et al. Single-cell RNA-sequencing analyses identify heterogeneity of CD8(+) T cell subpopulations and novel therapy targets in melanoma. Mol. Ther. Oncolytics 2020, 20, 105–118. [Google Scholar] [CrossRef]
- Kim, M.; Min, Y.K.; Jang, J.; Park, H.; Lee, S.; Lee, C.H. Single-cell RNA sequencing reveals distinct cellular factors for response to immunotherapy targeting CD73 and PD-1 in colorectal cancer. J. Immunother. Cancer 2021, 9, e002503. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef]
- Wu, L.; Mao, L.; Liu, J.-F.; Chen, L.; Yu, G.-T.; Yang, L.-L.; Wu, H.; Bu, L.-L.; Kulkarni, A.B.; Zhang, W.-F. Blockade of TIGIT/CD155 signaling reverses T-cell exhaustion and enhances antitumor capability in head and neck squamous cell carcinoma. Cancer Immunol. Res. 2019, 7, 1700–1713. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.L.; Maxwell, R.; Theodros, D.; Belcaid, Z.; Mathios, D.; Luksik, A.S.; Kim, E.; Wu, A.; Xia, Y.; Garzon-Muvdi, T.; et al. TIGIT and PD-1 dual checkpoint blockade enhances antitumor immunity and survival in GBM. Oncoimmunology 2018, 7, e1466769. [Google Scholar] [CrossRef] [PubMed]
- Sunseri, N.; Chen, X.; Wald, N.; Preillon, J.; Smith, S.M.; Driessens, G.; Kline, J. Beyond PD-1: Investigating the Therapeutic Potential of TIGIT Blockade in DLBCL; American Society of Hematology: Washington, DC, USA, 2019. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayden, J.A.; Côté, P.; Bombardier, C. Evaluation of the quality of prognosis studies in systematic reviews. Ann. Intern. Med. 2006, 144, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-L.; Wang, Y.-Y.; Yang, Z.-H.; Huang, D.; Weng, H.; Zeng, X.-T. Methodological quality (risk of bias) assessment tools for primary and secondary medical studies: What are they and which is better? Mil. Med. Res. 2020, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, R.J.; Thompson, S.G. A likelihood approach to meta-analysis with random effects. Stat. Med. 1996, 15, 619–629. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begg, C.B.; Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-J.; Zhou, Z.-Q.; Wang, P.; Chen, C.-L.; Liu, Y.; Pan, Q.-Z.; Zhu, Q.; Tang, Y.; Weng, D.-S.; Xia, J.-C. Orchestration of immune checkpoints in tumor immune contexture and their prognostic significance in esophageal squamous cell carcinoma. Cancer Manag. Res. 2018, 10, 6457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, X.; Wang, X.; Wang, T.; Zhang, X. Correlation of T cell immunoglobulin and ITIM Domain (TIGIT) and programmed death 1 (PD-1) with clinicopathological characteristics of renal cell carcinoma may indicate potential targets for treatment. Med Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 6861. [Google Scholar] [CrossRef]
- Tang, W.; Pan, X.; Han, D.; Rong, D.; Zhang, M.; Yang, L.; Ying, J.; Guan, H.; Chen, Z.; Wang, X. Clinical significance of CD8+ T cell immunoreceptor with Ig and ITIM domains+ in locally advanced gastric cancer treated with SOX regimen after D2 gastrectomy. Oncoimmunology 2019, 8, e1593807. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Lee, Y.J.; Choi, M.E.; Yun, K.A.; Won, C.H.; Lee, M.W.; Choi, J.H.; Chang, S.E. Expression of lymphocyte-activating gene 3 and T-cell immunoreceptor with immunoglobulin and ITIM domains in cutaneous melanoma and their correlation with programmed cell death 1 expression in tumor-infiltrating lymphocytes. J. Am. Acad. Dermatol. 2019, 81, 219–227. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, J.; Chen, Y.; Cui, J.; Lei, Y.; Cui, Y.; Jiang, N.; Jiang, W.; Chen, L.; Chen, Y. Combined evaluation of the expression status of CD155 and TIGIT plays an important role in the prognosis of LUAD (lung adenocarcinoma). Int. Immunopharmacol. 2020, 80, 106198. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, Q.; Wang, Z.; Zhang, H.; Zeng, H.; Huang, Q.; Chen, Y.; Jiang, W.; Lin, Z.; Qu, Y. Intratumoral TIGIT+ CD8+ T-cell infiltration determines poor prognosis and immune evasion in patients with muscle-invasive bladder cancer. J. Immunother. Cancer 2020, 8, e000978. [Google Scholar] [CrossRef]
- Shadbad, M.A.; Hajiasgharzadeh, K.; Derakhshani, A.; Silvestris, N.; Baghbanzadeh, A.; Racanelli, V.; Baradaran, B. From Melanoma Development to RNA-Modified Dendritic Cell Vaccines: Highlighting the Lessons From the Past. Front. Immunol. 2021, 12, 331. [Google Scholar] [CrossRef]
- Stanietsky, N.; Rovis, T.L.; Glasner, A.; Seidel, E.; Tsukerman, P.; Yamin, R.; Enk, J.; Jonjic, S.; Mandelboim, O. Mouse TIGIT inhibits NK-cell cytotoxicity upon interaction with PVR. Eur. J. Immunol. 2013, 43, 2138–2150. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Li, L.; Lu, F.; Yue, J.; Liu, Z.; Zhang, W.; Fu, R. Overexpression of TIGIT in NK and T Cells Contributes to Tumor Immune Escape in Myelodysplastic Syndromes. Front. Oncol. 2020, 10, 1595. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, H.; Han, F.; Chen, X.; Lin, R.; Wang, W.; Qiu, H.; Zhuang, Z.; Liao, Q.; Zhang, W. CD155T/TIGIT signaling regulates CD8+ T-cell metabolism and promotes tumor progression in human gastric cancer. Cancer Res. 2017, 77, 6375–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.-M.; Li, W.-Q.; Wu, Y.-H.; Han, L.; Cao, X.-G.; Yang, X.-M.; Wang, H.-F.; Zhao, W.-S.; Zhai, W.-J.; Qi, Y.-M. Intrinsic expression of immune checkpoint molecule TIGIT could help tumor growth in vivo by suppressing the function of NK and CD8+ T cells. Front. Immunol. 2018, 9, 2821. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yu, Y.; Yang, S.; Zeng, B.; Zhang, Z.; Jiao, G.; Zhang, Y.; Cai, L.; Yang, R. Regulation of arginase I activity and expression by both PD-1 and CTLA-4 on the myeloid-derived suppressor cells. Cancer Immunol. Immunother. 2009, 58, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Finn, O.J.; Ochoa, A.C. myeloid derived suppressor cells as disease modulators. Front. Immunol. 2020, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Dufait, I.; Schwarze, J.K.; Liechtenstein, T.; Leonard, W.; Jiang, H.; Escors, D.; De Ridder, M.; Breckpot, K. Ex vivo generation of myeloid-derived suppressor cells that model the tumor immunosuppressive environment in colorectal cancer. Oncotarget 2015, 6, 12369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuvers, M.E.; Muskens, F.; Bezemer, K.; Lambers, M.; Dingemans, A.-M.C.; Groen, H.J.; Smit, E.F.; Hoogsteden, H.C.; Hegmans, J.P.; Aerts, J.G. Arginase-1 mRNA expression correlates with myeloid-derived suppressor cell levels in peripheral blood of NSCLC patients. Lung Cancer 2013, 81, 468–474. [Google Scholar] [CrossRef]
- Ren, W.; Zhang, X.; Li, W.; Feng, Q.; Feng, H.; Tong, Y.; Rong, H.; Wang, W.; Zhang, D.; Zhang, Z. Circulating and tumor-infiltrating arginase 1-expressing cells in gastric adenocarcinoma patients were mainly immature and monocytic Myeloid-derived suppressor cells. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Pang, X.; Fan, H.-Y.; Tang, Y.-L.; Wang, S.-S.; Cao, M.-X.; Wang, H.-F.; Dai, L.-L.; Wang, K.; Yu, X.-H.; Wu, J.-B. Myeloid derived suppressor cells contribute to the malignant progression of oral squamous cell carcinoma. PLoS ONE 2020, 15, e0229089. [Google Scholar] [CrossRef] [Green Version]
- Han, H.S.; Jeong, S.; Kim, H.; Kim, H.-D.; Kim, A.R.; Kwon, M.; Park, S.-H.; Woo, C.G.; Kim, H.K.; Lee, K.H. TOX-expressing terminally exhausted tumor-infiltrating CD8+ T cells are reinvigorated by co-blockade of PD-1 and TIGIT in bladder cancer. Cancer Lett. 2020, 499, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Inozume, T.; Yaguchi, T.; Furuta, J.; Harada, K.; Kawakami, Y.; Shimada, S. Melanoma cells control antimelanoma CTL responses via interaction between TIGIT and CD155 in the effector phase. J. Investig. Dermatol. 2016, 136, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Abreu, D.; Johnson, M.L.; Hussein, M.A.; Cobo, M.; Patel, A.J.; Secen, N.M.; Lee, K.H.; Massuti, B.; Hiret, S.; Yang, J.C.-H. Primary Analysis of a Randomized, Double-Blind, Phase II Study of the Anti-TIGIT Antibody Tiragolumab (tira) Plus Atezolizumab (atezo) versus Placebo Plus Atezo as First-Line (1L) Treatment in Patients with PD-L1-Selected NSCLC (CITYSCAPE); American Society of Clinical Oncology: Alexandria, VA, USA, 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).