Regional Differences in the Permeability Barrier of the Skin—Implications in Acantholytic Skin Diseases
Abstract
:1. Introduction
2. Results
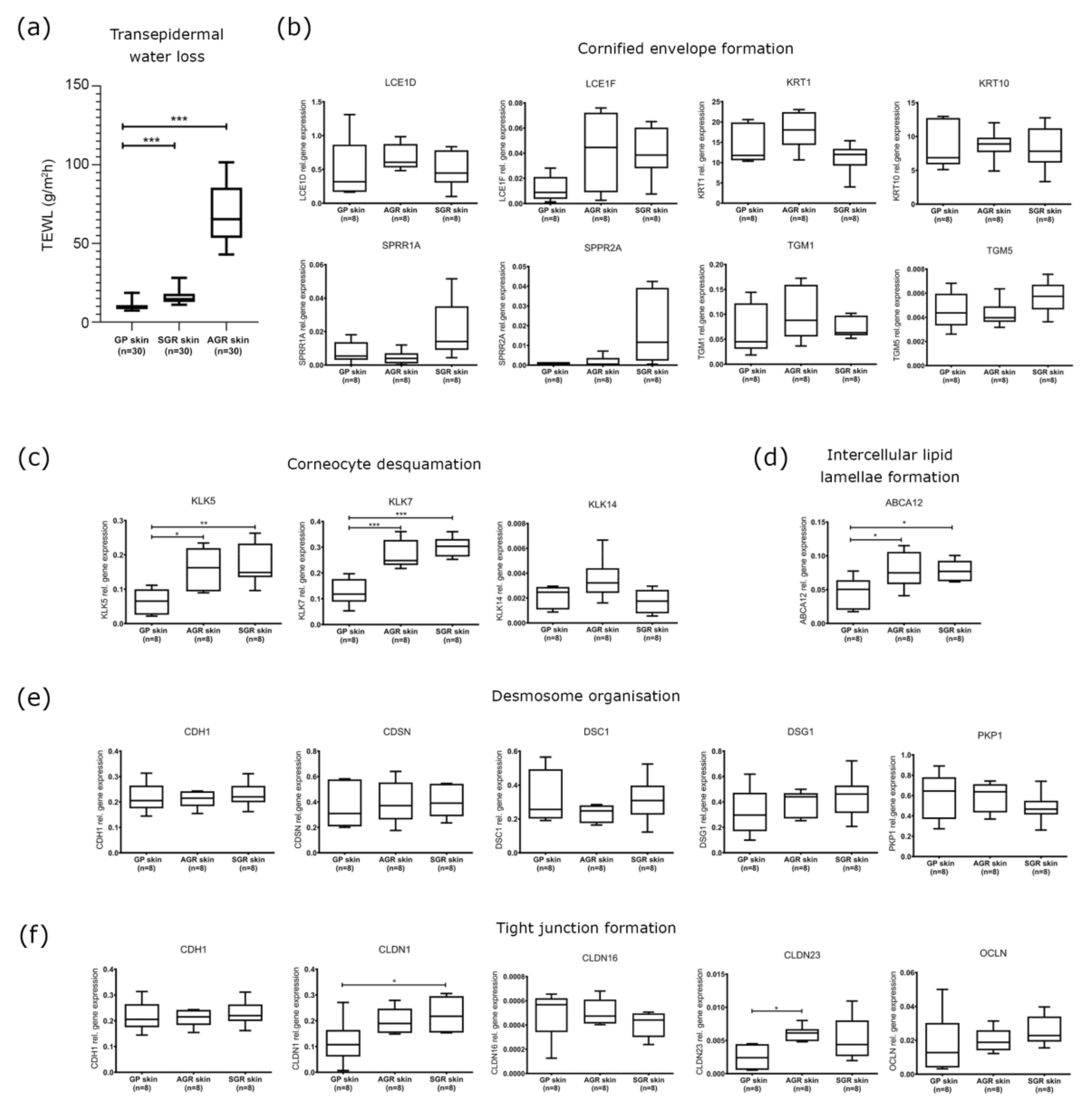
2.1. Distinct Barrier Functions Characterize Different Skin Regions
2.2. Cornified Envelope Intracellular Structure Molecules Similarly Expressed in Distinct Skin Areas
2.3. Increased Expression of KLK Proteases and the ABCA12 Lipid Transporter Characterize SGR and AGR Skin Regions
2.4. Decreased Protein Expression of (Corneo)Desmosome and Tight Junction Components in SGR and AGR Skin Regions
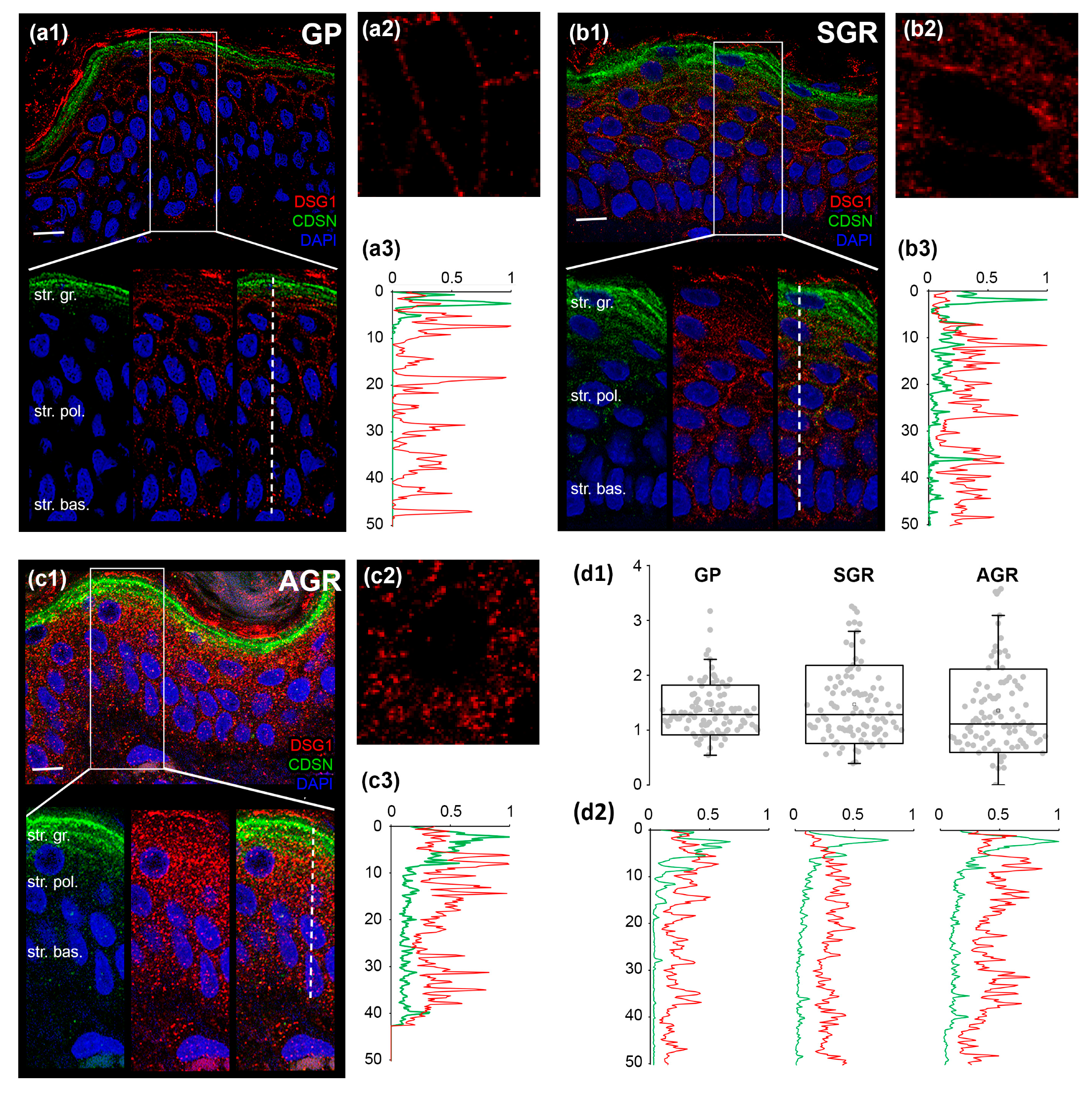
2.5. Distinct Organization of Cell Junctions in the Permeability Barrier of Different Regions
3. Discussion
4. Materials and Methods
4.1. Measurement of Transepidermal Water Loss
4.2. Skin Biopsies
4.3. Processing of Skin Biopsy Specimens
4.4. RNA Isolation, Reverse Transcription
4.5. Real-Time Polymerase Chain Reaction (RT-qPCR)
4.6. Immunohistochemistry
4.7. Confocal Microscopy
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Christensen, G.J.M.; Nakatsuji, T.; et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef] [Green Version]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Genet. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Jenei, A.; Dajnoki, Z.; Medgyesi, B.; Gáspár, K.; Béke, G.; Kinyó, Á.; Méhes, G.; Hendrik, Z.; Dinya, T.; Törőcsik, D.; et al. Apocrine Gland–Rich Skin Has a Non-Inflammatory IL-17–Related Immune Milieu, that Turns to Inflammatory IL-17–Mediated Disease in Hidradenitis Suppurativa. J. Investig. Dermatol. 2019, 139, 964–968. [Google Scholar] [CrossRef] [Green Version]
- Béke, G.; Dajnoki, Z.; Kapitány, A.; Gáspár, K.; Medgyesi, B.; Póliska, S.; Hendrik, Z.; Péter, Z.; Törőcsik, D.; Bíró, T.; et al. Immunotopographical Differences of Human Skin. Front. Immunol. 2018, 9, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dajnoki, Z.; Béke, G.; Kapitány, A.; Mócsai, G.; Gáspár, K.; Rühl, R.; Hendrik, Z.; Juhász, I.; Zouboulis, C.C.; Bacsi, A.; et al. Sebaceous Gland-Rich Skin Is Characterized by TSLP Expression and Distinct Immune Surveillance Which Is Disturbed in Rosacea. J. Investig. Dermatol. 2017, 137, 1114–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borkowski, A.W.; Gallo, R.L. The Coordinated Response of the Physical and Antimicrobial Peptide Barriers of the Skin. J. Investig. Dermatol. 2011, 131, 285–287. [Google Scholar] [CrossRef] [Green Version]
- Dorschner, R.; Pestonjamasp, V.K.; Tamakuwala, S.; Ohtake, T.; Rudisill, J.; Nizet, V.; Agerberth, B.; Gudmundsson, G.H.; Gallo, R. Cutaneous Injury Induces the Release of Cathelicidin Anti-Microbial Peptides Active Against Group A Streptococcus. J. Investig. Dermatol. 2001, 117, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauber, J.; Gallo, R.L. Expanding the Roles of Antimicrobial Peptides in Skin: Alarming and Arming Keratinocytes. J. Investig. Dermatol. 2007, 127, 510–512. [Google Scholar] [CrossRef] [Green Version]
- Aberg, K.M.; Man, M.-Q.; Gallo, R.L.; Ganz, T.; Crumrine, D.; Brown, B.E.; Choi, E.-H.; Kim, D.-K.; Schröder, J.M.; Feingold, K.R.; et al. Co-Regulation and Interdependence of the Mammalian Epidermal Permeability and Antimicrobial Barriers. J. Investig. Dermatol. 2008, 128, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Egawa, G.; Kabashima, K. Barrier dysfunction in the skin allergy. Allergol. Int. 2018, 67, 3–11. [Google Scholar] [CrossRef]
- Egawa, G.; Kabashima, K. Multifactorial skin barrier deficiency and atopic dermatitis: Essential topics to prevent the atopic march. J. Allergy Clin. Immunol. 2016, 138, 350–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGovern, J.; Meinert, C.; De Veer, S.; Hollier, B.; Parker, T.; Upton, Z.; De Veer, S. Attenuated kallikrein-related peptidase activity disrupts desquamation and leads to stratum corneum thickening in human skin equivalent models. Br. J. Dermatol. 2016, 176, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Kishibe, M. Physiological and pathological roles of kallikrein-related peptidases in the epidermis. J. Dermatol. Sci. 2019, 95, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Ishida-Yamamoto, A.; Kishibe, M. Involvement of corneodesmosome degradation and lamellar granule transportation in the desquamation process. Med. Mol. Morphol. 2011, 44, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Kleesz, P.; Darlenski, R.; Fluhr, J. Full-Body Skin Mapping for Six Biophysical Parameters: Baseline Values at 16 Anatomical Sites in 125 Human Subjects. Ski. Pharmacol. Physiol. 2012, 25, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, N.; Tsai, B.; Sidiropoulos, M.; Saijoh, K.; Levesque, M.A.; Takehara, K.; Diamandis, E. Quantification of Eight Tissue Kallikreins in the Stratum Corneum and Sweat. J. Investig. Dermatol. 2006, 126, 925–929. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, N.; Saijoh, K.; Toyama, T.; Ohka, R.; Otsuki, N.; Hussack, G.; Takehara, K.; Diamandis, E. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br. J. Dermatol. 2005, 153, 274–281. [Google Scholar] [CrossRef]
- Stefansson, K.; Brattsand, M.; Roosterman, D.; Kempkes, C.; Bocheva, G.; Steinhoff, M.; Egelrud, T. Activation of Proteinase-Activated Receptor-2 by Human Kallikrein-Related Peptidases. J. Investig. Dermatol. 2008, 128, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Nadeau, P.; Henehan, M.; De Benedetto, A. Activation of protease-activated receptor 2 leads to impairment of keratinocyte tight junction integrity. J. Allergy Clin. Immunol. 2018, 142, 281–284.e7. [Google Scholar] [CrossRef] [Green Version]
- Henehan, M.; De Benedetto, A. Update on protease-activated receptor 2 in cutaneous barrier, differentiation, tumorigenesis and pigmentation, and its role in related dermatologic diseases. Exp. Dermatol. 2019, 28, 877–885. [Google Scholar] [CrossRef] [Green Version]
- Koziel, J.; Potempa, J. Protease-armed bacteria in the skin. Cell Tissue Res. 2012, 351, 325–337. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhang, Z.-W.; Luo, Y.; Liang, N.; Pi, X.-X.; Fan, Y.-M. Molecular epidemiology, in vitro susceptibility and exoenzyme screening of Malassezia clinical isolates. J. Med. Microbiol. 2020, 69, 436–442. [Google Scholar] [CrossRef]
- White, T.; Findley, K.; Dawson, T.; Scheynius, A.; Boekhout, T.; Cuomo, C.; Xu, J.; Saunders, C.W. Fungi on the Skin: Dermatophytes and Malassezia. Cold Spring Harb. Perspect. Med. 2014, 4, a019802. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.H.; Myrold, D.D.; Mueller, R.S. Distributions of Extracellular Peptidases Across Prokaryotic Genomes Reflect Phylogeny and Habitat. Front. Microbiol. 2019, 10, 413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dessinioti, C.; Katsambas, A.D. The role of Propionibacterium acnes in acne pathogenesis: Facts and controversies. Clin. Dermatol. 2010, 28, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.R.; Nakatsuji, T.; Sanford, J.; Vrbanac, A.F.; Gallo, R.L. Staphylococcus aureus Induces Increased Serine Protease Activity in Keratinocytes. J. Investig. Dermatol. 2016, 137, 377–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirasawa, Y.; Takai, T.; Nakamura, T.; Mitsuishi, K.; Gunawan, H.; Suto, H.; Ogawa, T.; Wang, X.-L.; Ikeda, S.; Okumura, K.; et al. Staphylococcus aureus Extracellular Protease Causes Epidermal Barrier Dysfunction. J. Investig. Dermatol. 2009, 130, 614–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujiie, I.; Ujiie, H.; Iwata, H.; Shimizu, H. Clinical and immunological features of pemphigus relapse. Br. J. Dermatol. 2019, 180, 1498–1505. [Google Scholar] [CrossRef]
- Schmidt, E.; Kasperkiewicz, M.; Joly, P. Pemphigus. Lancet 2019, 394, 882–894. [Google Scholar] [CrossRef]
- Kasperkiewicz, M.; Ellebrecht, C.T.; Takahashi, H.; Yamagami, J.; Zillikens, D.; Payne, A.S.; Amagai, M. Pemphigus. Nat. Rev. Dis. Primers 2017, 3, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savignac, M.; Simon, M.; Edir, A.; Guibbal, L.; Hovnanian, A. SERCA2 Dysfunction in Darier Disease Causes Endoplasmic Reticulum Stress and Impaired Cell-to-Cell Adhesion Strength: Rescue by Miglustat. J. Investig. Dermatol. 2014, 134, 1961–1970. [Google Scholar] [CrossRef] [Green Version]
- Dhitavat, J.; Fairclough, R.; Hovnanian, A.; Burge, S. Calcium pumps and keratinocytes: Lessons from Darier’s disease and Hailey-Hailey disease. Br. J. Dermatol. 2004, 150, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Sudbrak, R.; Brown, J.M.; Dobson-Stone, C.; Carter, S.; Ramser, J.; White, J.; Healy, E.; Dissanayake, M.; Larrègue, M.; Perrussel, M.; et al. Hailey-Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca2+ pump. Hum. Mol. Genet. 2000, 9, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Savignac, M.; Edir, A.; Simon, M.; Hovnanian, A. Darier disease: A disease model of impaired calcium homeostasis in the skin. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1813, 1111–1117. [Google Scholar] [CrossRef] [Green Version]



| Healthy Individuals (n = 8) | |||
|---|---|---|---|
| Healthy individuals | Sex | Age | Localization |
| GP1 | F | 43 | upper arm |
| GP2 | F | 66 | ankle |
| GP3 | F | 50 | upper arm |
| GP4 | F | 52 | shin |
| GP5 | M | 25 | upper arm |
| GP6 | M | 50 | shin |
| GP7 | M | 65 | upper arm |
| GP8 | M | 67 | ankle |
| Mean age ± SD | 52.2 ± 14.1 | ||
| Healthy Individuals (n = 8) | |||
| Healthy individuals | Sex | Age | Localization |
| AGR1 | M | 70 | axilla |
| AGR2 | M | 43 | axilla |
| AGR3 | F | 48 | axilla |
| AGR4 | F | 65 | axilla |
| AGR5 | F | 60 | axilla |
| AGR6 | F | 38 | axilla |
| AGR7 | F | 45 | axilla |
| AGR8 | F | 41 | axilla |
| Mean age ± SD | 51.2 ± 12.0 | ||
| Healthy Individuals (n = 8) | |||
| Healthy individuals | Sex | Age | Localization |
| SGR1 | M | 42 | nose |
| SGR2 | M | 61 | forehead |
| SGR3 | F | 62 | chin |
| SGR4 | M | 66 | nose |
| SGR5 | M | 38 | face |
| SGR6 | F | 69 | forehead |
| SGR7 | M | 68 | nose |
| SGR8 | M | 67 | face |
| Mean age ± SD | 59.1 ± 12.2 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapitány, A.; Medgyesi, B.; Jenei, A.; Somogyi, O.; Szabó, L.; Gáspár, K.; Méhes, G.; Hendrik, Z.; Dócs, K.; Szücs, P.; et al. Regional Differences in the Permeability Barrier of the Skin—Implications in Acantholytic Skin Diseases. Int. J. Mol. Sci. 2021, 22, 10428. https://doi.org/10.3390/ijms221910428
Kapitány A, Medgyesi B, Jenei A, Somogyi O, Szabó L, Gáspár K, Méhes G, Hendrik Z, Dócs K, Szücs P, et al. Regional Differences in the Permeability Barrier of the Skin—Implications in Acantholytic Skin Diseases. International Journal of Molecular Sciences. 2021; 22(19):10428. https://doi.org/10.3390/ijms221910428
Chicago/Turabian StyleKapitány, Anikó, Barbara Medgyesi, Adrienn Jenei, Orsolya Somogyi, Lilla Szabó, Krisztián Gáspár, Gábor Méhes, Zoltán Hendrik, Klaudia Dócs, Péter Szücs, and et al. 2021. "Regional Differences in the Permeability Barrier of the Skin—Implications in Acantholytic Skin Diseases" International Journal of Molecular Sciences 22, no. 19: 10428. https://doi.org/10.3390/ijms221910428
APA StyleKapitány, A., Medgyesi, B., Jenei, A., Somogyi, O., Szabó, L., Gáspár, K., Méhes, G., Hendrik, Z., Dócs, K., Szücs, P., Dajnoki, Z., & Szegedi, A. (2021). Regional Differences in the Permeability Barrier of the Skin—Implications in Acantholytic Skin Diseases. International Journal of Molecular Sciences, 22(19), 10428. https://doi.org/10.3390/ijms221910428







