Lineage-Specific Variation in IR Boundary Shift Events, Inversions, and Substitution Rates among Caprifoliaceae s.l. (Dipsacales) Plastomes
Abstract
:1. Introduction
2. Results
2.1. Plastome Organization
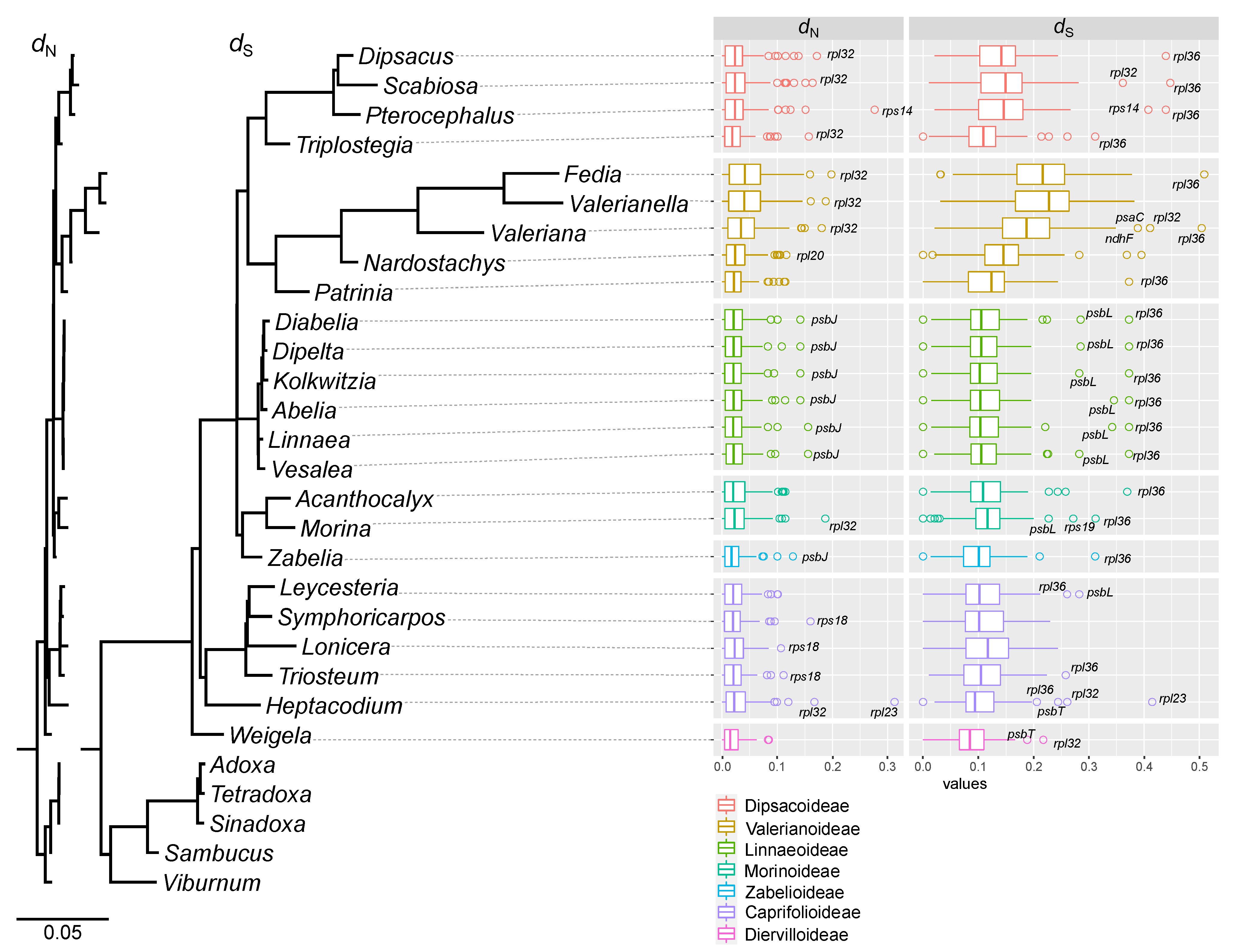
2.2. Elevated Substitution Rates in the Plastomes of the Subfamily Valerianoideae
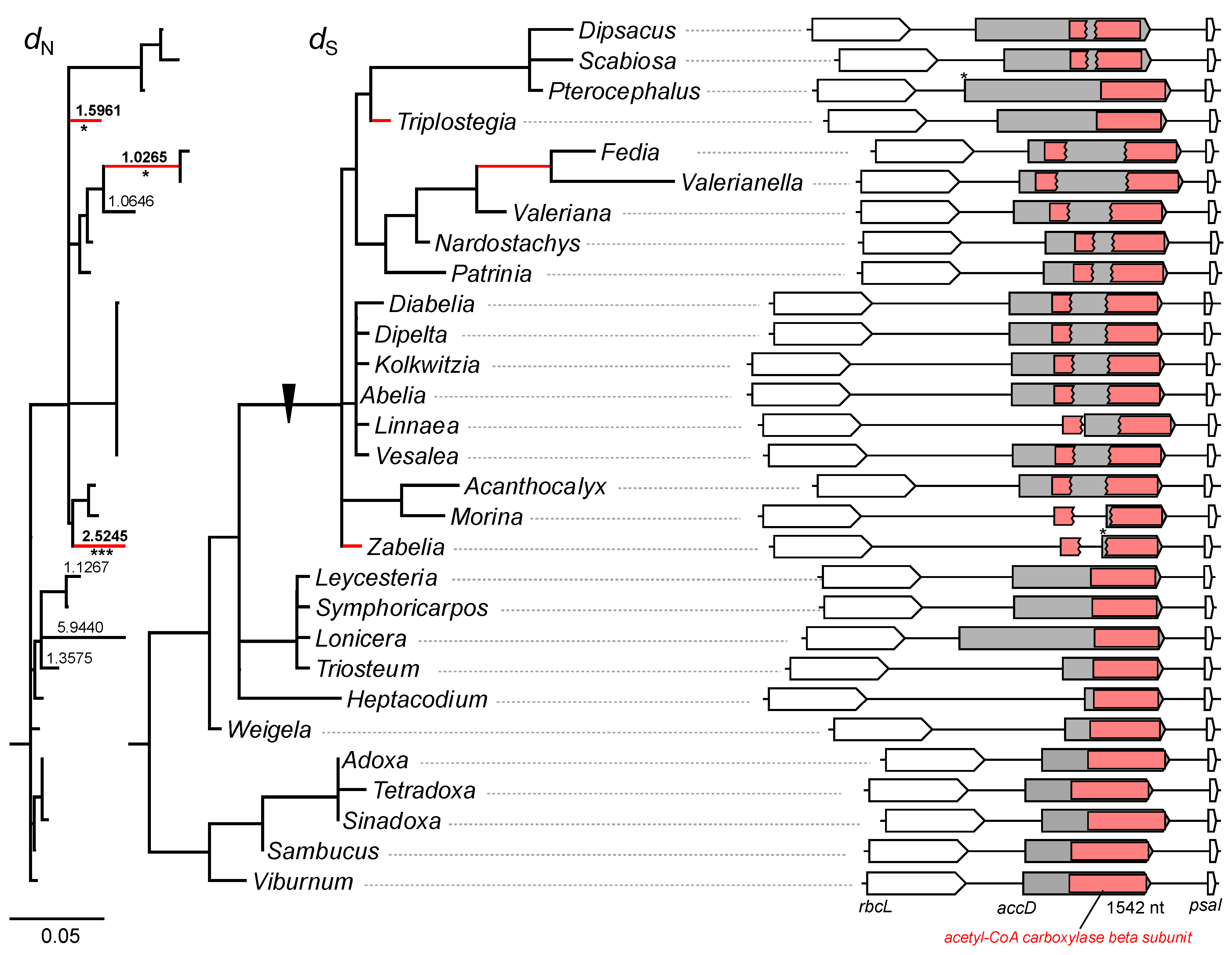
2.3. Structural Evolution of Plastid-Encoded accD Gene in Caprifoliaceae s.l. Plastomes
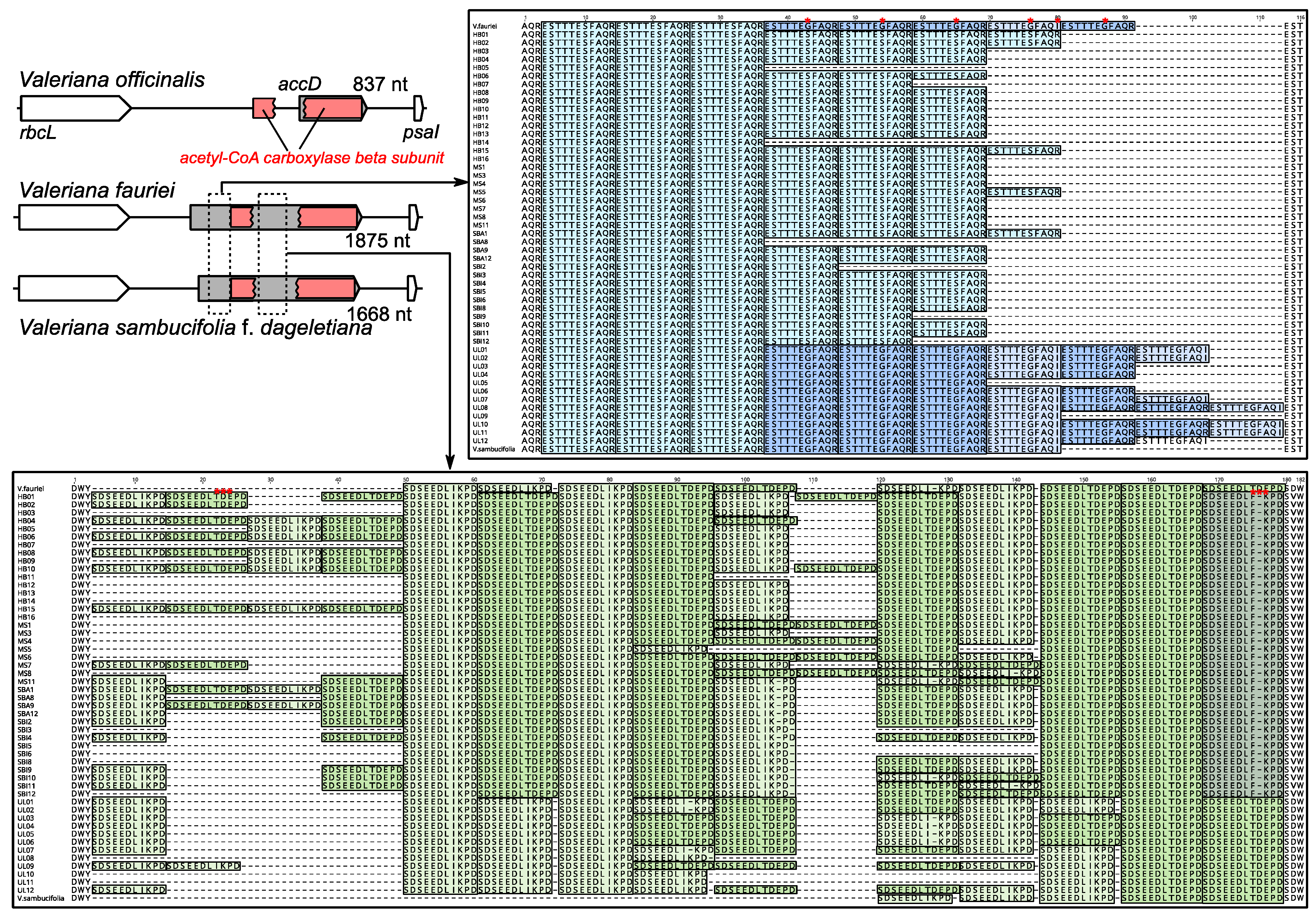
2.4. Length Variation in the Valeriana accD Gene
2.5. Correlation between Structural Change and Substitution Rates in the Plastid-Encoded clpP Gene
3. Discussion
4. Materials and Methods
4.1. Genome Sequencing, Assembly, and Annotation
4.2. Comparative Analyses
4.3. Estimation of Substitution Rates
4.4. Survey of Variability in the Plastid-Encoded accD Gene
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruhlman, T.A.; Jansen, R.K. The plastid genomes of flowering plants. Methods Mol. Biol. 2014, 1132, 3–38. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Cosner, M.E.; Raubeson, L.A.; Jansen, R.K. Chloroplast DNA rearrangements in Campanulaceae: Phylogenetic utility of highly rearranged genomes. BMC Evol. Biol. 2004, 4, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, D.B.; Triant, D.A.; Forrester, N.J.; Bergner, L.M.; Wu, M.; Taylor, D.R. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol. Phylogenetics Evol. 2014, 72, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Guisinger, M.; Kim, H.-G.; Ruck, E.; Blazier, J.C.; McMurtry, V.; Kuehl, J.V.; Boore, J.; Jansen, R.K. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 2008, 67, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.-L.; Jansen, R.K.; Chumley, T.W.; Kim, K.-J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; An, B.; Park, S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 2018, 8, 13568. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Wang, D. Comparative Chloroplast Genomics of Corydalis Species (Papaveraceae): Evolutionary Perspectives on Their Unusual Large Scale Rearrangements. Front. Plant Sci. 2021, 11. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. USA 2008, 105, 18424–18429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, E.N.; Ruhlman, T.A.; Weng, M.-L.; Khiyami, M.A.; Sabir, J.S.M.; Hajarah, N.H.; Alharbi, N.S.; Rabah, S.O.; Jansen, R.K. Plastome-Wide Nucleotide Substitution Rates Reveal Accelerated Rates in Papilionoideae and Correlations with Genome Features Across Legume Subfamilies. J. Mol. Evol. 2017, 84, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Weng, M.-L.; Theriot, E.C.; Gilbert, L.E.; Ruhlman, T.A.; Krosnick, S.E.; Jansen, R.K. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol. Phylogenetics Evol. 2019, 138, 53–64. [Google Scholar] [CrossRef]
- Weng, M.-L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Ruhlman, T.A.; Weng, M.-L.; Hajrah, N.H.; Sabir, J.S.M.; Jansen, R.K. Contrasting Patterns of Nucleotide Substitution Rates Provide Insight into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium. Genome Biol. Evol. 2017, 9, 1766–1780. [Google Scholar] [CrossRef]
- Magee, A.M.; Aspinall, S.; Rice, D.W.; Cusack, B.P.; Sémon, M.; Perry, A.S.; Stefanović, S.; Milbourne, D.; Barth, S.; Palmer, J.D.; et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010, 20, 1700–1710. [Google Scholar] [CrossRef] [Green Version]
- Blazier, J.C.; Ruhlman, T.A.; Weng, M.-L.; Rehman, S.K.; Sabir, J.S.M.; Jansen, R.K. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci. Rep. 2016, 6, 24595. [Google Scholar] [CrossRef] [Green Version]
- Barnard-Kubow, K.B.; Sloan, D.B.; Galloway, L.F. Correlation between sequence divergence and polymorphism reveals similar evolutionary mechanisms acting across multiple timescales in a rapidly evolving plastid genome. BMC Evol. Biol. 2014, 14, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkinson, C.L.; Mower, J.P.; Qiu, Y.-L.; Shirk, A.J.; Song, K.; Young, N.D.; Claude, W.d.; Palmer, J.D. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol. Biol. 2005, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Rousseau-Gueutin, M.; Huang, X.; Higginson, E.; Ayliffe, M.; Day, A.; Timmis, J.N. Potential Functional Replacement of the Plastidic Acetyl-CoA Carboxylase Subunit (accD) Gene by Recent Transfers to the Nucleus in Some Angiosperm Lineages. Plant Physiol. 2013, 161, 1918–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, J.; Hazzouri, K.M.; Nickrent, D.; Geisler, M.; Meyer, R.S.; Pentony, M.M.; Flowers, J.M.; Pelser, P.; Barcelona, J.; Inovejas, S.A.; et al. Possible Loss of the Chloroplast Genome in the Parasitic Flowering Plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 2014, 31, 793–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabir, J.; Schwarz, E.; Ellison, N.; Zhang, J.; Baeshen, N.A.; Mutwakil, M.; Jansen, R.; Ruhlman, T. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 2014, 12, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Babiychuk, E.; Vandepoele, K.; Wissing, J.; Garcia-Diaz, M.; De Rycke, R.; Akbari, H.; Joubès, J.; Beeckman, T.; Jänsch, L.; Frentzen, M.; et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 2011, 108, 6674–6679. [Google Scholar] [CrossRef] [Green Version]
- Schulte, W.; Töpfer, R.; Stracke, R.; Schell, J.; Martini, N. Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: Indication for plastidic localization of at least one isoform. Proc. Natl. Acad. Sci. USA 1997, 94, 3465–3470. [Google Scholar] [CrossRef] [Green Version]
- Konishi, T.; Shinohara, K.; Yamada, K.; Sasaki, Y. Acetyl-CoA Carboxylase in Higher Plants: Most Plants Other Than Gramineae Have Both the Prokaryotic and the Eukaryotic Forms of This Enzyme. Plant Cell Physiol. 1996, 37, 117–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gornicki, P.; Faris, J.; King, I.; Podkowinski, J.; Gill, B.; Haselkorn, R. Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc. Natl. Acad. Sci. USA 1997, 94, 14179–14184. [Google Scholar] [CrossRef] [Green Version]
- Rockenbach, K.; Havird, J.C.; Monroe, J.G.; Triant, D.A.; Taylor, D.R.; Sloan, D.B. Positive Selection in Rapidly Evolving Plastid–Nuclear Enzyme Complexes. Genetics 2016, 204, 1507–1522. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.M.; Friso, G.; van Wijk, K.J.; Sloan, D.B. Extreme variation in rates of evolution in the plastid Clp protease complex. Plant J. 2019, 98, 243–259. [Google Scholar] [CrossRef]
- White, S.W.; Zheng, J.; Zhang, Y.M.; Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Wakil, S.J.; Stoops, J.K.; Joshi, V.C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983, 52, 537–579. [Google Scholar] [CrossRef]
- Adam, Z.; Adamska, I.; Nakabayashi, K.; Ostersetzer, O.; Haussuhl, K.; Manuell, A.; Zheng, B.; Vallon, O.; Rodermel, S.R.; Shinozaki, K.; et al. Chloroplast and Mitochondrial Proteases in Arabidopsis. A Proposed Nomenclature1. Plant Physiol. 2001, 125, 1912–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikanai, T.; Shimizu, K.; Ueda, K.; Nishimura, Y.; Kuroiwa, T.; Hashimoto, T. The Chloroplast clpP Gene, Encoding a Proteolytic Subunit of ATP-Dependent Protease, is Indispensable for Chloroplast Development in Tobacco. Plant Cell Physiol. 2001, 42, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-X.; Liu, H.; Moore, M.J.; Landrein, S.; Liu, B.; Zhu, Z.-X.; Wang, H.-F. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales). Mol. Phylogenetics Evol. 2020, 142, 106641. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-X.; Morales-Briones, D.F.; Moore, M.J.; Wen, J.; Wang, H.-F. A phylogenomic perspective on gene tree conflict and character evolution in Caprifoliaceae using target enrichment data, with Zabelioideae recognized as a new subfamily. J. Syst. Evol. 2021, 59, 897–914. [Google Scholar] [CrossRef]
- Fan, W.-B.; Wu, Y.; Yang, J.; Shahzad, K.; Li, Z.-H. Comparative Chloroplast Genomics of Dipsacales Species: Insights Into Sequence Variation, Adaptive Evolution, and Phylogenetic Relationships. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Goulding, S.E.; Wolfe, K.; Olmstead, R.; Morden, C. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. MGG 1996, 252, 195–206. [Google Scholar] [CrossRef]
- Haberle, R.C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008, 66, 350–361. [Google Scholar] [CrossRef]
- Gemayel, R.; Vinces, M.D.; Legendre, M.; Verstrepen, K.J. Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences. Annu. Rev. Genet. 2010, 44, 445–477. [Google Scholar] [CrossRef] [Green Version]
- Gurdon, C.; Maliga, P. Two Distinct Plastid Genome Configurations and Unprecedented Intraspecies Length Variation in the accD Coding Region in Medicago truncatula. DNA Res. 2014, 21, 417–427. [Google Scholar] [CrossRef] [Green Version]
- Erixon, P.; Oxelman, B. Whole-Gene Positive Selection, Elevated Synonymous Substitution Rates, Duplication, and Indel Evolution of the Chloroplast clpP1 Gene. PLoS ONE 2008, 3, e1386. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-C.; Chen, S.-Y.; Zhang, X.-Z. Chloroplast Genome Evolution in Actinidiaceae: clpP Loss, Heterogenous Divergence and Phylogenomic Practice. PLoS ONE 2016, 11, e0162324. [Google Scholar] [CrossRef]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- Hepburn, N.J.; Schmidt, D.W.; Mower, J.P. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol. Biol. Evol. 2012, 29, 3111–3120. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Kosakovsky Pond, S.L.; Poon, A.F.Y.; Velazquez, R.; Weaver, S.; Hepler, N.L.; Murrell, B.; Shank, S.D.; Magalis, B.R.; Bouvier, D.; Nekrutenko, A.; et al. HyPhy 2.5—A Customizable Platform for Evolutionary Hypothesis Testing Using Phylogenies. Mol. Biol. Evol. 2019, 37, 295–299. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Kosakovsky Pond, S.L.; Scheffler, K. RELAX: Detecting Relaxed Selection in a Phylogenetic Framework. Mol. Biol. Evol. 2014, 32, 820–832. [Google Scholar] [CrossRef] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, G.; Flores-Vergara, M.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Taxon | Dipsacoideae | Valerianoideae | |||
|---|---|---|---|---|---|
| Dipsacus japonicus | Scabiosa comosa | Fedia cornucopiae | Valeriana fauriei | Valerianella locusta | |
| Size (bp) | 160,243 | 159,651 | 152,196 | 155,302 | 149,809 |
| LSC length (bp) | 87,066 | 87,477 | 82,960 | 85,541 | 82,103 |
| SSC length (bp) | 17,850 | 18,716 | 15,862 | 15,159 | 15,796 |
| IR length (bp) | 27,664 | 26,729 | 26,687 | 27,301 | 25,955 |
| Number of protein-coding genes | 78 (6) | 78 (7) | 79 (4) | 79 (4) | 79 (4) |
| Number of tRNA genes | 29 (8) | 30 (7) | 30 (7) | 30 (7) | 30 (7) |
| Number of rRNA genes | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| Number of introns | 21 (5) | 20 (5) | 21 (5) | 21 (5) | 19 (5) |
| GC content (%) | 38.8 | 38.7 | 38.2 | 38.4 | 38.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Jun, M.; Park, S.; Park, S. Lineage-Specific Variation in IR Boundary Shift Events, Inversions, and Substitution Rates among Caprifoliaceae s.l. (Dipsacales) Plastomes. Int. J. Mol. Sci. 2021, 22, 10485. https://doi.org/10.3390/ijms221910485
Park S, Jun M, Park S, Park S. Lineage-Specific Variation in IR Boundary Shift Events, Inversions, and Substitution Rates among Caprifoliaceae s.l. (Dipsacales) Plastomes. International Journal of Molecular Sciences. 2021; 22(19):10485. https://doi.org/10.3390/ijms221910485
Chicago/Turabian StylePark, Seongjun, Minji Jun, Sunmi Park, and SeonJoo Park. 2021. "Lineage-Specific Variation in IR Boundary Shift Events, Inversions, and Substitution Rates among Caprifoliaceae s.l. (Dipsacales) Plastomes" International Journal of Molecular Sciences 22, no. 19: 10485. https://doi.org/10.3390/ijms221910485






