Autophagy-Related Proteins Are Differentially Expressed in Adrenal Cortical Tumor/Pheochromocytoma and Associated with Patient Prognosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. In Silico Analysis
2.3. Tissue Microarray
2.4. Immunohistochemistry
2.5. Interpretation of Immunohistochemical Staining
2.6. Statistical Analysis
3. Results
3.1. Clinicopathologic Characteristics of Patients
3.2. Expression of Autophagy-Related Proteins in ACT and PCC
3.3. Presence/Absence of Clinicopathologic Factors in PCC/ACT and Expression of Autophagy-Related Proteins
3.4. Impact of the Expression of Autophagy-Related Proteins on Patient Prognosis in PCC/ACT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Klionsky, D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010, 12, 814–822. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lu, Y.; Lu, C.; Zhang, L. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (hif)-1alpha expression. Pathol. Oncol. Res. 2009, 15, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Li, B.X.; Li, C.Y.; Peng, R.Q.; Wu, X.J.; Wang, H.Y.; Wan, D.S.; Zhu, X.F.; Zhang, X.S. The expression of beclin 1 is associated with favorable prognosis in stage iiib colon cancers. Autophagy 2009, 5, 303–306. [Google Scholar] [CrossRef] [Green Version]
- Pirtoli, L.; Cevenini, G.; Tini, P.; Vannini, M.; Oliveri, G.; Marsili, S.; Mourmouras, V.; Rubino, G.; Miracco, C. The prognostic role of beclin 1 protein expression in high-grade gliomas. Autophagy 2009, 5, 930–936. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.B.; Fan, X.J.; Chen, M.Y.; Xiang, J.; Huang, P.Y.; Guo, L.; Wu, X.Y.; Xu, J.; Long, Z.J.; Zhao, Y.; et al. Elevated beclin 1 expression is correlated with hif-1alpha in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy 2010, 6, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. Lc3, a mammalian homologue of yeast apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Sivridis, E.; Koukourakis, M.I.; Zois, C.E.; Ledaki, I.; Ferguson, D.J.; Harris, A.L.; Gatter, K.C.; Giatromanolaki, A. Lc3a-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am. J. Pathol. 2010, 176, 2477–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, A.; Miyata, H.; Doki, Y.; Yamasaki, M.; Sohma, I.; Gotoh, K.; Takiguchi, S.; Fujiwara, Y.; Uchiyama, Y.; Monden, M. Lc3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int. J. Oncol. 2008, 33, 461–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinsay, M.N.; Thomas, R.L.; Lee, Y.; Gustafsson, A.B. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy 2010, 6, 855–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gelinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Debnath, J. Autophagy and tumorigenesis. Semin. Immunopathol. 2010, 32, 383–396. [Google Scholar] [CrossRef] [Green Version]
- Baehrecke, E.H. Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef]
- Debnath, J.; Baehrecke, E.H.; Kroemer, G. Does autophagy contribute to cell death? Autophagy 2005, 1, 66–74. [Google Scholar] [CrossRef]
- Henry, L.R.; Lee, H.O.; Lee, J.S.; Klein-Szanto, A.; Watts, P.; Ross, E.A.; Chen, W.T.; Cheng, J.D. Clinical implications of fibroblast activation protein in patients with colon cancer. Clin. cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 1736–1741. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.; Pfeifer, U.; Dämmrich, J. Inhibited autophagic degradation during acth-stimulated growth of rat adrenal zona fasciculata. Virchows Archiv. B 1986, 52, 429. [Google Scholar] [CrossRef]
- Yi, J.; Tang, X. Functional implication of autophagy in steroid-secreting cells of the rat. Anat. Rec. 1995, 242, 137–146. [Google Scholar] [CrossRef]
- Cheon, S.Y.; Kim, H.; Rubinsztein, D.C.; Lee, J.E. Autophagy, cellular aging and age-related human diseases. Exp. Neurobiol. 2019, 28, 643–657. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Thorburn, A.; Debnath, J. Autophagy and metastasis: Another double-edged sword. Curr. Opin. Cell Biol. 2010, 22, 241–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosa, M.S.; Bragado, P.; Aguirre-Ghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.R.; Fidler, I.J. The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 2011, 128, 2527–2535. [Google Scholar] [CrossRef] [Green Version]
- Weckman, A.; Rotondo, F.; Di Ieva, A.; Syro, L.V.; Butz, H.; Cusimano, M.D.; Kovacs, K. Autophagy in endocrine tumors. Endocr. Relat. Cancer 2015, 22, R205–R218. [Google Scholar] [CrossRef] [Green Version]
- Maiti, A.; Hait, N.C. Autophagy-mediated tumor cell survival and progression of breast cancer metastasis to the brain. J. Cancer 2021, 12, 954–964. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, Y.; Zou, J.; Ouyang, J.; Cai, Z.; Zeng, X.; Ling, H.; Zeng, T. Autophagy and its role in gastric cancer. Clin. Chim. Acta 2019, 489, 10–20. [Google Scholar] [CrossRef]
- Thongchot, S.; Vidoni, C.; Ferraresi, A.; Loilome, W.; Khuntikeo, N.; Sangkhamanon, S.; Titapun, A.; Isidoro, C.; Namwat, N. Cancer-associated fibroblast-derived il-6 determines unfavorable prognosis in cholangiocarcinoma by affecting autophagy-associated chemoresponse. Cancers (Basel) 2021, 13, 2134. [Google Scholar] [CrossRef]
- Fabrizi, C.; De Vito, S.; Somma, F.; Pompili, E.; Catizone, A.; Leone, S.; Lenzi, P.; Fornai, F.; Fumagalli, L. Lithium improves survival of pc12 pheochromocytoma cells in high-density cultures and after exposure to toxic compounds. Int. J. Cell Biol. 2014, 2014, 135908. [Google Scholar] [CrossRef] [Green Version]
- Amaravadi, R.K.; Yu, D.; Lum, J.J.; Bui, T.; Christophorou, M.A.; Evan, G.I.; Thomas-Tikhonenko, A.; Thompson, C.B. Autophagy inhibition enhances therapy-induced apoptosis in a myc-induced model of lymphoma. J. Clin. Investig. 2007, 117, 326–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carew, J.S.; Medina, E.C.; Esquivel, J.A., 2nd; Mahalingam, D.; Swords, R.; Kelly, K.; Zhang, H.; Huang, P.; Mita, A.C.; Mita, M.M.; et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J. Cell Mol. Med. 2010, 14, 2448–2459. [Google Scholar] [CrossRef] [PubMed]
- Carew, J.S.; Nawrocki, S.T.; Kahue, C.N.; Zhang, H.; Yang, C.; Chung, L.; Houghton, J.A.; Huang, P.; Giles, F.J.; Cleveland, J.L. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor saha to overcome bcr-abl-mediated drug resistance. Blood 2007, 110, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Roy, S.; Lazar, A.J.; Wang, W.L.; McAuliffe, J.C.; Reynoso, D.; McMahon, J.; Taguchi, T.; Floris, G.; Debiec-Rychter, M.; et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (gist). Proc. Natl. Acad. Sci. USA 2010, 107, 14333–14338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerquetti, L.; Sampaoli, C.; Amendola, D.; Bucci, B.; Masuelli, L.; Marchese, R.; Misiti, S.; De Venanzi, A.; Poggi, M.; Toscano, V.; et al. Rosiglitazone induces autophagy in h295r and cell cycle deregulation in sw13 adrenocortical cancer cells. Exp. Cell Res. 2011, 317, 1397–1410. [Google Scholar] [CrossRef]
- Ikeda, T.; Ishii, K.A.; Saito, Y.; Miura, M.; Otagiri, A.; Kawakami, Y.; Shimano, H.; Hara, H.; Takekoshi, K. Inhibition of autophagy enhances sunitinib-induced cytotoxicity in rat pheochromocytoma pc12 cells. J. Pharmacol. Sci. 2013, 121, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Tanaka, Y.; Aita, Y.; Ishii, K.; Ikeda, T.; Isobe, K.; Kawakami, Y.; Shimano, H.; Hara, H.; Takekoshi, K. Sunitinib induces apoptosis in pheochromocytoma tumor cells by inhibiting vegfr2/akt/mtor/s6k1 pathways through modulation of bcl-2 and bad. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E615–E625. [Google Scholar] [CrossRef] [Green Version]
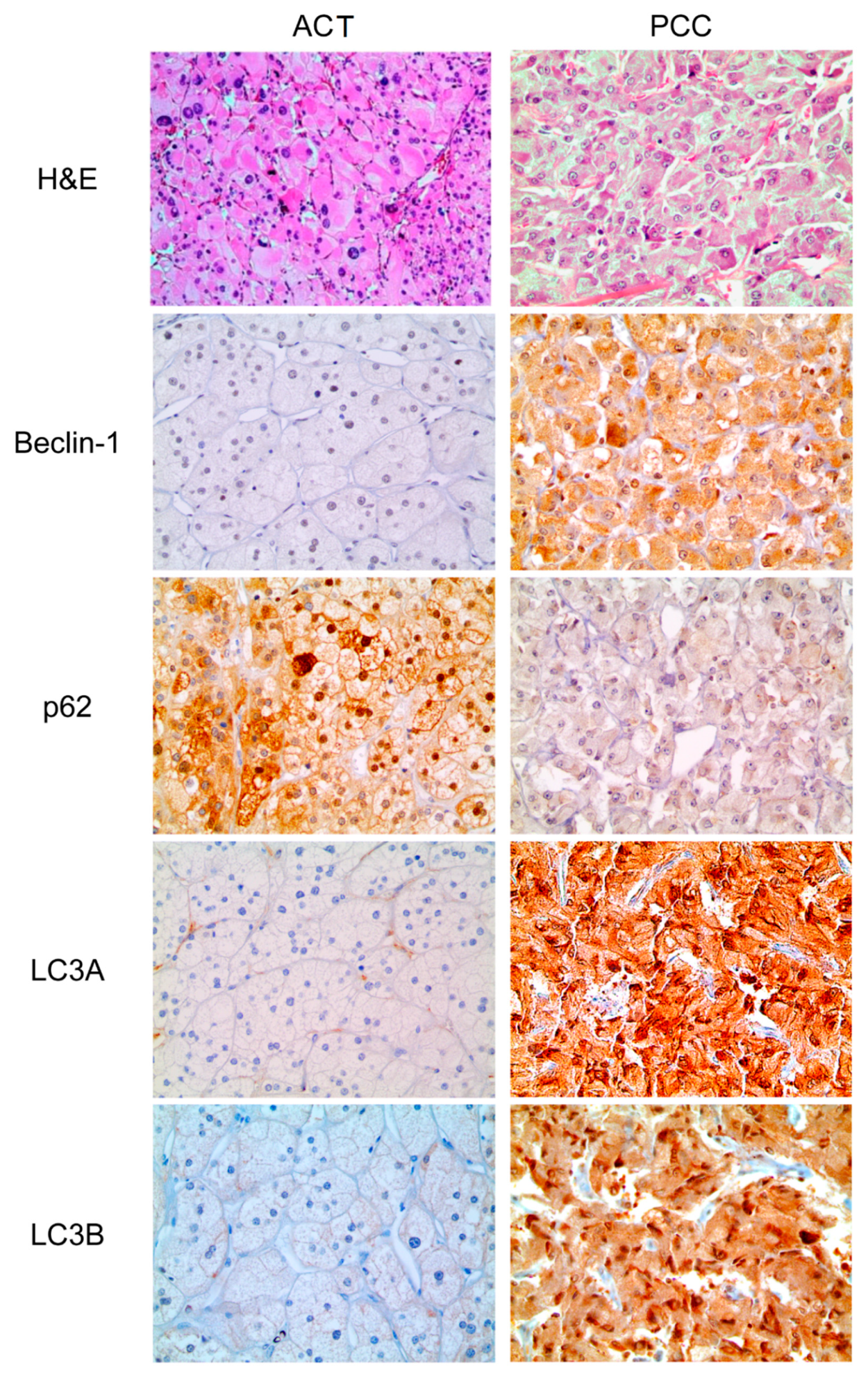
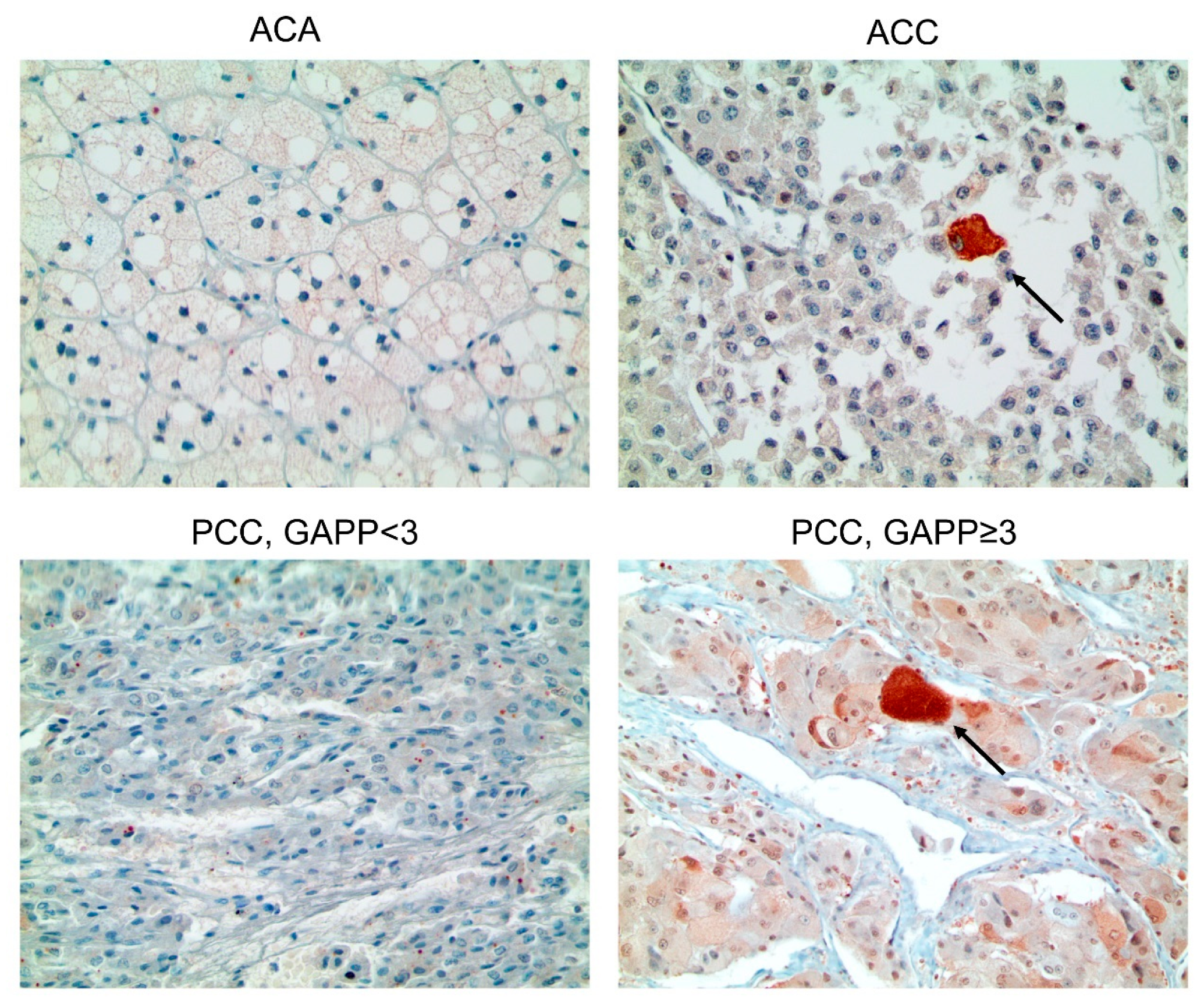
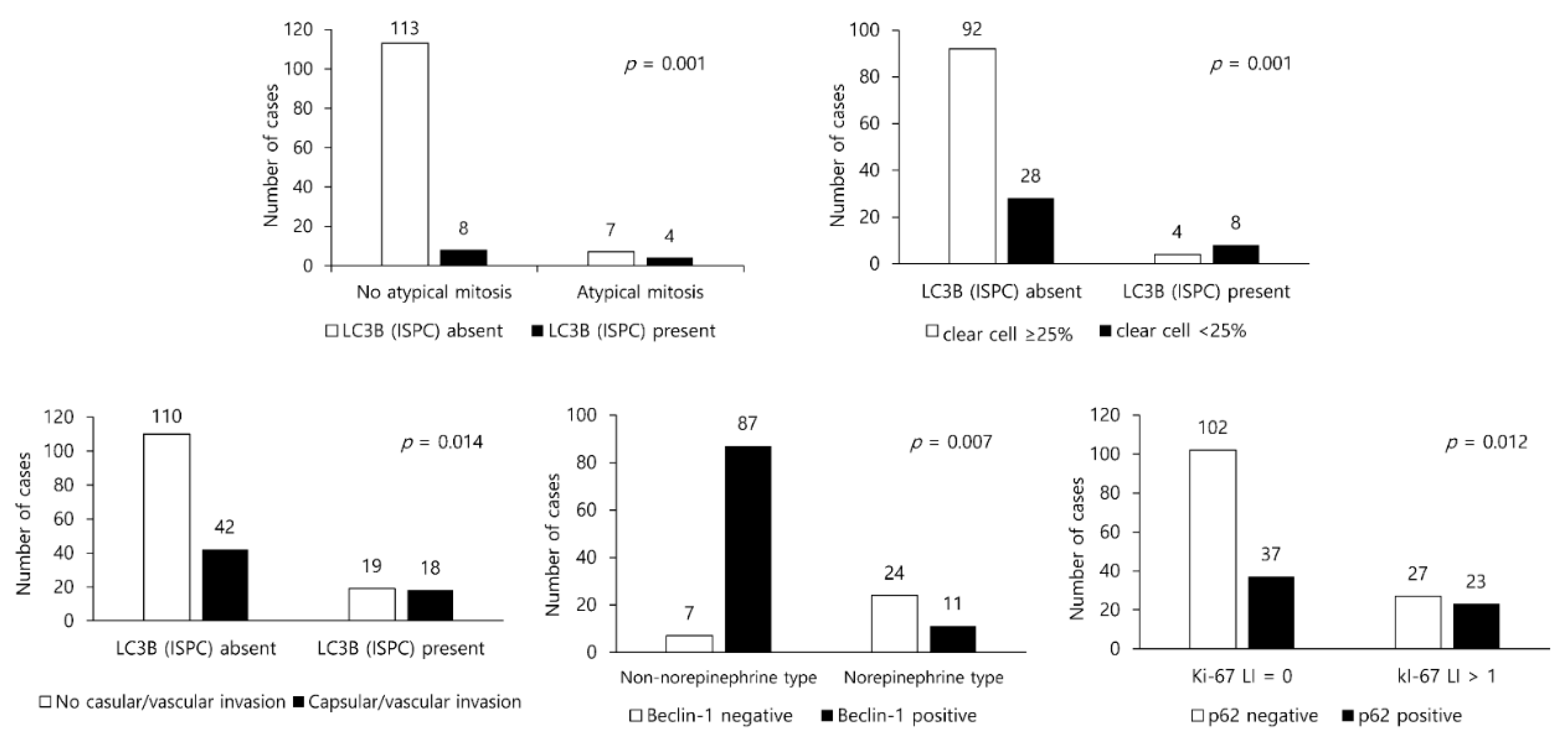

| Parameters | Total n = 321 (%) | Adrenal Cortical Tumor n = 132 (%) | Pheochromocytoma n = 189 (%) | p-Value |
|---|---|---|---|---|
| Beclin-1 | <0.001 | |||
| Negative | 218 (67.9) | 127 (96.2) | 91 (48.1) | |
| Positive | 103 (32.1) | 5 (3.8) | 98 (51.9) | |
| p62 | 0.006 | |||
| Negative | 199 (62.0) | 70 (53.0) | 129 (68.3) | |
| Positive | 122 (38.0) | 62 (47.0) | 60 (31.7) | |
| p62 (ISPC) | 0.128 | |||
| Absent | 238 (74.1) | 92 (69.7) | 146 (77.2) | |
| Present | 83 (25.9) | 40 (30.3) | 43 (22.8) | |
| LC3A | <0.001 | |||
| Negative | 221 (68.8) | 132 (100.0) | 89 (47.1) | |
| Positive | 100 (31.2) | 0 (0.0) | 100 (52.9) | |
| LC3B | <0.001 | |||
| Negative | 243 (75.7) | 116 (87.9) | 127 (67.2) | |
| Positive | 78 (24.3) | 16 (12.1) | 62 (32.8) | |
| LC3B (ISPC) | 0.010 | |||
| Absent | 272 (84.7) | 120 (90.9) | 152 (80.4) | |
| Present | 49 (15.3) | 12 (9.1) | 37 (19.6) |
| Parameters | Total n = 132 (%) | Adrenal Cortical Adenoma n = 115 (%) | Adrenal Cortical Carcinoma n = 17 (%) | p-Value |
|---|---|---|---|---|
| Beclin-1 | 0.381 | |||
| Negative | 127 (96.2) | 110 (95.7) | 17 (100.0) | |
| Positive | 5 (3.8) | 5 (4.3) | 0 (0.0) | |
| p62 | 0.294 | |||
| Negative | 70 (53.0) | 63 (54.8) | 7 (41.2) | |
| Positive | 62 (47.0) | 52 (45.2) | 10 (58.8) | |
| p62 (ISPC) | 0.107 | |||
| Absent | 92 (69.7) | 83 (72.2) | 9 (52.9) | |
| Present | 40 (30.3) | 32 (27.8) | 8 (47.1) | |
| LC3A | n/a | |||
| Negative | 132 (100.0) | 115 (100.0) | 17 (100.0) | |
| Positive | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| LC3B | 0.101 | |||
| Negative | 116 (87.9) | 99 (86.1) | 17 (100.0) | |
| Positive | 16 (12.1) | 16 (13.9) | 0 (0.0) | |
| LC3B (ISPC) | 0.027 | |||
| Absent | 120 (90.9) | 107 (93.0) | 13 (76.5) | |
| Present | 12 (9.1) | 8 (7.0) | 4 (23.5) |
| Parameters | Total n = 189 (%) | Pheochromocytoma | p-Value | |
|---|---|---|---|---|
| GAPP Score < 3 n = 138 (%) | GAPP Score ≥ 3 n = 51 (%) | |||
| Beclin-1 | 0.884 | |||
| Negative | 91 (48.1) | 66 (47.8) | 25 (49.0) | |
| Positive | 98 (51.9) | 72 (52.2) | 26 (51.0) | |
| p62 | 0.017 | |||
| Negative | 129 (68.3) | 101 (73.2) | 28 (54.9) | |
| Positive | 60 (31.7) | 37 (26.8) | 23 (45.1) | |
| p62 (ISPC) | 0.349 | |||
| Absent | 146 (77.2) | 109 (79.0) | 37 (72.5) | |
| Present | 43 (22.8) | 29 (21.0) | 14 (27.5) | |
| LC3A | 0.747 | |||
| Negative | 89 (47.1) | 64 (46.4) | 25 (49.0) | |
| Positive | 100 (52.9) | 74 (53.6) | 26 (51.0) | |
| LC3B | 0.254 | |||
| Negative | 127 (67.2) | 96 (69.6) | 31 (60.8) | |
| Positive | 62 (32.8) | 42 (30.4) | 20 (39.2) | |
| LC3B (ISPC) | 0.013 | |||
| Absent | 152 (80.4) | 117 (84.8) | 35 (68.6) | |
| Present | 37 (19.6) | 21 (15.2) | 16 (31.4) | |
| Parameter | Number of Patients /Recurrence/Death | Disease-Free Survival | Overall Survival | ||
|---|---|---|---|---|---|
| Mean Survival (95% CI) Months | p-Value | Mean Survival (95% CI) Months | p-Value | ||
| Beclin-1 | 0.655 | 0.112 | |||
| Negative | 91/2/8 | 150 (139–161) | 142 (126–158) | ||
| Positive | 97/3/3 | 154 (148–161) | 159 (150–167) | ||
| p62 | 0.014 | 0.023 | |||
| Negative | 128/1/4 | 156 (154–159) | 150 (142–157) | ||
| Positive | 60/4/7 | 139 (115–162) | 136 (114–157) | ||
| p62 (ISPC) | 0.001 | 0.910 | |||
| Absent | 145/1/9 | 159 (157–162) | 144 (134–155) | ||
| Present | 43/4/2 | 127 (96–157) | 157 (144–170) | ||
| LC3A | 0.657 | 0.435 | |||
| Negative | 89/3/4 | 146 (136–157) | 154 (142–166) | ||
| Positive | 99/2/7 | 157 (151–162) | 142 (129–155) | ||
| LC3B | 0.529 | 0.220 | |||
| Negative | 126/4/5 | 149 (140–158) | 155 (145–165) | ||
| Positive | 62/1/6 | 157 (151–164) | 138 (121–155) | ||
| LC3B (ISPC) | 0.234 | 0.410 | |||
| Absent | 151/3/10 | 154 (147–162) | 148 (137–160) | ||
| Present | 37/2/1 | 145 (129–162) | 151 (139–163) | ||
| Included Factor | Disease-Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Histologic pattern | 0.738 | 0.720 | ||||
| Zellballen vs. Non-Zellballen | 1.491 | 0.143–15,351 | 0.737 | 0.139–3.912 | ||
| Cellularity | 0.290 | 0.504 | ||||
| Low, moderate vs. High | 3.093 | 0.382–25.02 | 1.827 | 0.312–10.71 | ||
| Vascular and/or capsular invasion | 0.524 | 0.183 | ||||
| Absent vs. Present | 2.125 | 0.210–21.55 | 2.854 | 0.610–13.34 | ||
| Ki-67 labeling index (%) | 0.923 | 0.112 | ||||
| <1 vs. ≥1 | 1.598 | 0.000–21,497 | 0.124 | 0.010–1.623 | ||
| GAPP score | 0.892 | 0.061 | ||||
| 0–2 vs. 3–10 | 1.949 | 0.000–29,027 | 13.906 | 0.884–218.8 | ||
| p62 | 0.420 | 0.015 | ||||
| Negative vs. Positive | 2.945 | 0.123–40.72 | 6.240 | 1.434–27.15 | ||
| p62 (ISPC) | 0.081 | 0.432 | ||||
| Absent vs. Present | 8.143 | 0.771–85.95 | 0.522 | 0.103–2.638 | ||
| Parameter | Number of Patients /Recurrence/Death | Disease-Free Survival | Overall Survival | ||
|---|---|---|---|---|---|
| Mean Survival (95% CI) Months | p-Value | Mean Survival (95% CI) Months | p-Value | ||
| Beclin-1 | n/a | n/a | |||
| Negative | 127/3/9 | n/a | n/a | ||
| Positive | 5/0/0 | n/a | n/a | ||
| p62 | 0.446 | 0.056 | |||
| Negative | 70/1/2 | 107 (104–110) | 106 (102–110) | ||
| Positive | 62/2/7 | 115 (110–120) | 106 (97–115) | ||
| p62 (ISPC) | 0.138 | 0.322 | |||
| Absent | 92/1/5 | 107 (105–109) | 103 (99–108) | ||
| Present | 40/2/4 | 113 (105–121) | 107 (96–118) | ||
| LC3A | n/a | n/a | |||
| Negative | 132/3/9 | n/a | n/a | ||
| Positive | 0/0/0 | n/a | n/a | ||
| LC3B | n/a | n/a | |||
| Negative | 116/3/9 | n/a | n/a | ||
| Positive | 16/0/0 | n/a | n/a | ||
| LC3B (ISPC) | 0.094 | 0.157 | |||
| Absent | 120/2/7 | 107 (105–109) | 103 (98–107) | ||
| Present | 12/1/2 | 108 (88–128) | 100 (77–123) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.M.; Koo, J.S. Autophagy-Related Proteins Are Differentially Expressed in Adrenal Cortical Tumor/Pheochromocytoma and Associated with Patient Prognosis. Int. J. Mol. Sci. 2021, 22, 10490. https://doi.org/10.3390/ijms221910490
Kim HM, Koo JS. Autophagy-Related Proteins Are Differentially Expressed in Adrenal Cortical Tumor/Pheochromocytoma and Associated with Patient Prognosis. International Journal of Molecular Sciences. 2021; 22(19):10490. https://doi.org/10.3390/ijms221910490
Chicago/Turabian StyleKim, Hye Min, and Ja Seung Koo. 2021. "Autophagy-Related Proteins Are Differentially Expressed in Adrenal Cortical Tumor/Pheochromocytoma and Associated with Patient Prognosis" International Journal of Molecular Sciences 22, no. 19: 10490. https://doi.org/10.3390/ijms221910490
APA StyleKim, H. M., & Koo, J. S. (2021). Autophagy-Related Proteins Are Differentially Expressed in Adrenal Cortical Tumor/Pheochromocytoma and Associated with Patient Prognosis. International Journal of Molecular Sciences, 22(19), 10490. https://doi.org/10.3390/ijms221910490






