Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders
Abstract
:1. Introduction
2. Results
2.1. Even and Odd Medium Chain-Fatty Acids (mc-FAs) Induce Different Change in the Cellular Lipidome of Long-Chain Fatty Acid Oxidation Disorders (lc-FAOD)
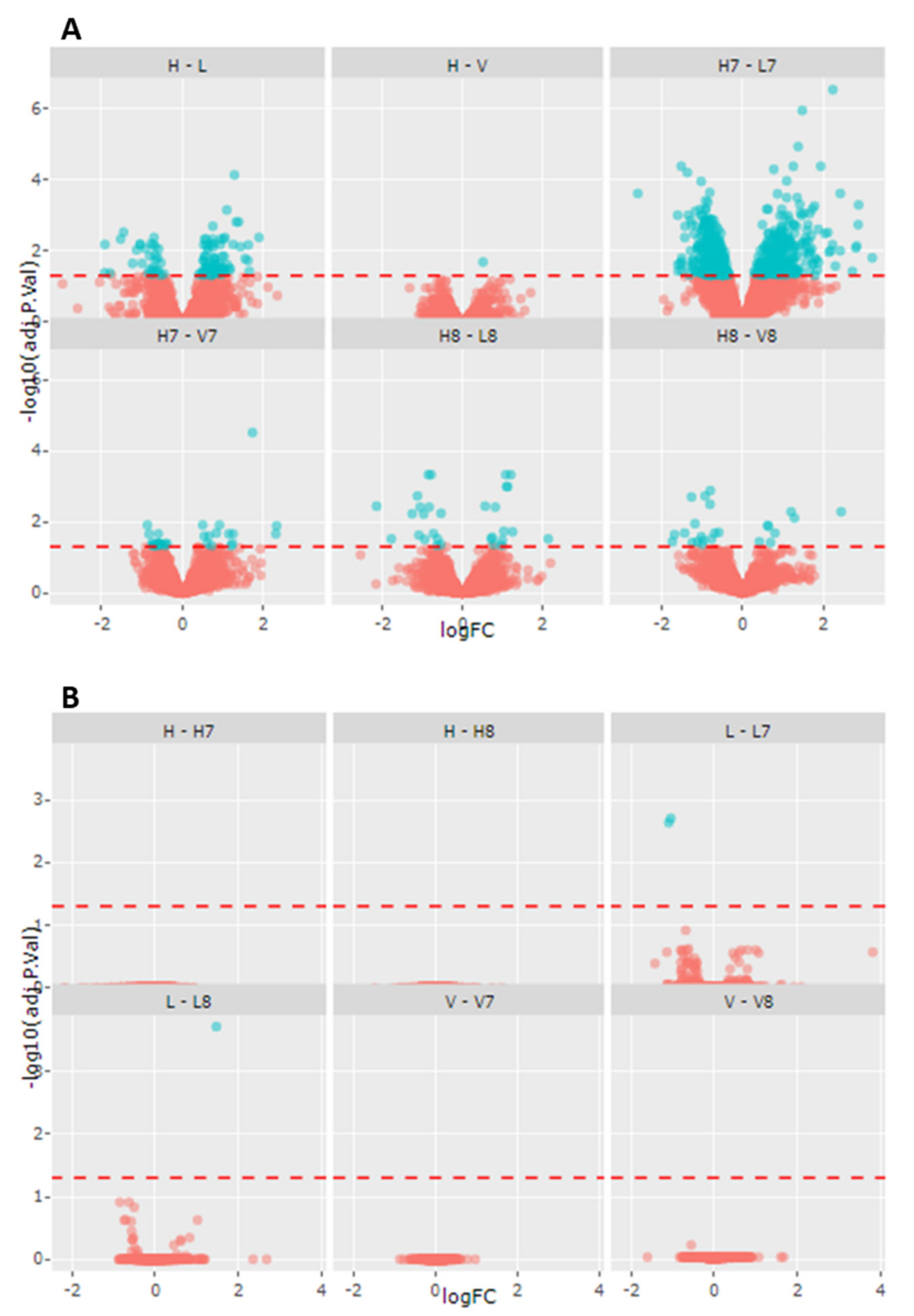
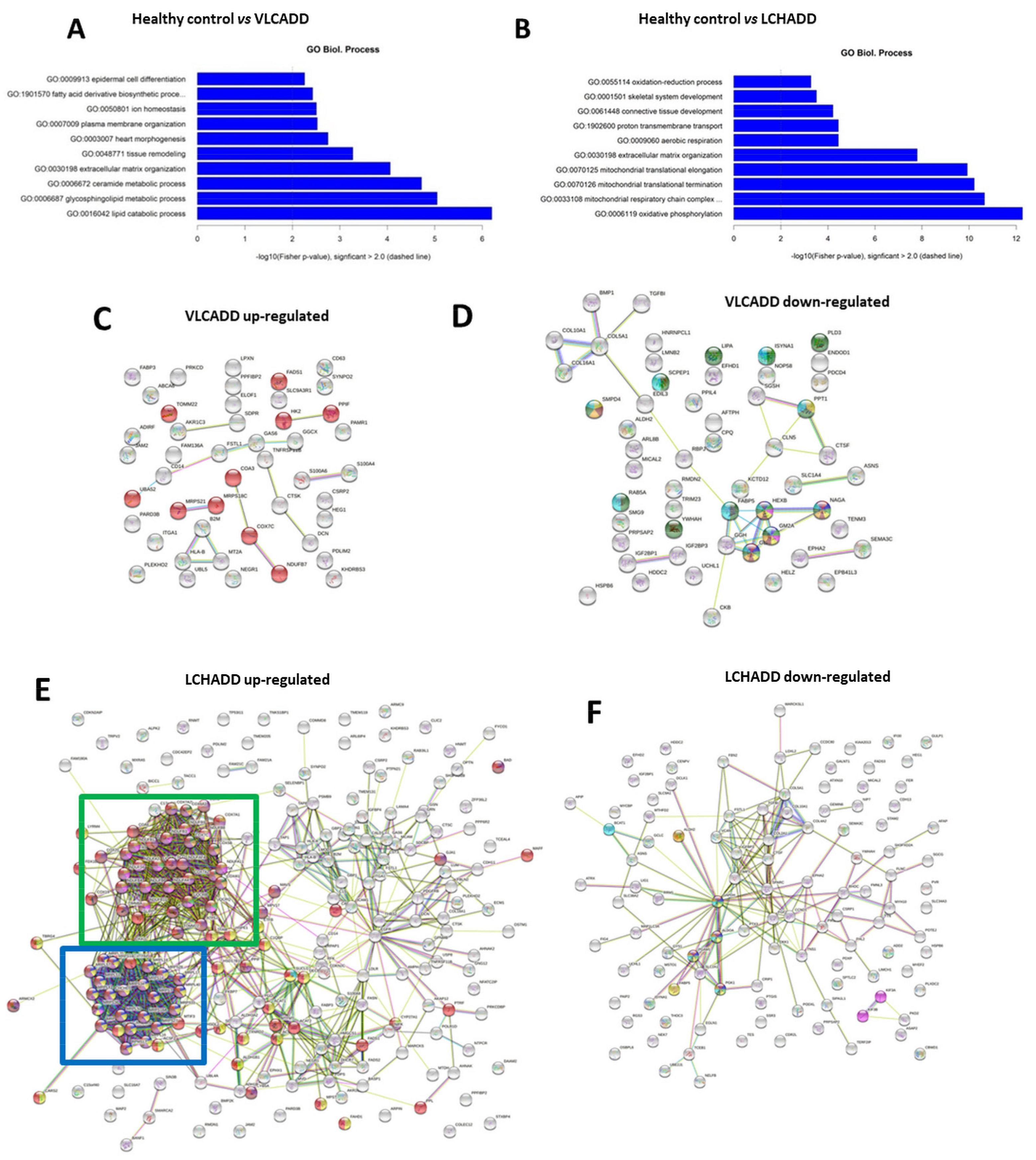
2.2. Gentotype and Not Mc-FAs Induce Disease Specific Proteome Aleration in Long-Chain Fatty Acid Oxidation Disorders (lc-FAOD)
2.3. Mc-FAs Deeply Alter Composition and Desaturation Degree of Mitochondrial Cardiolipins
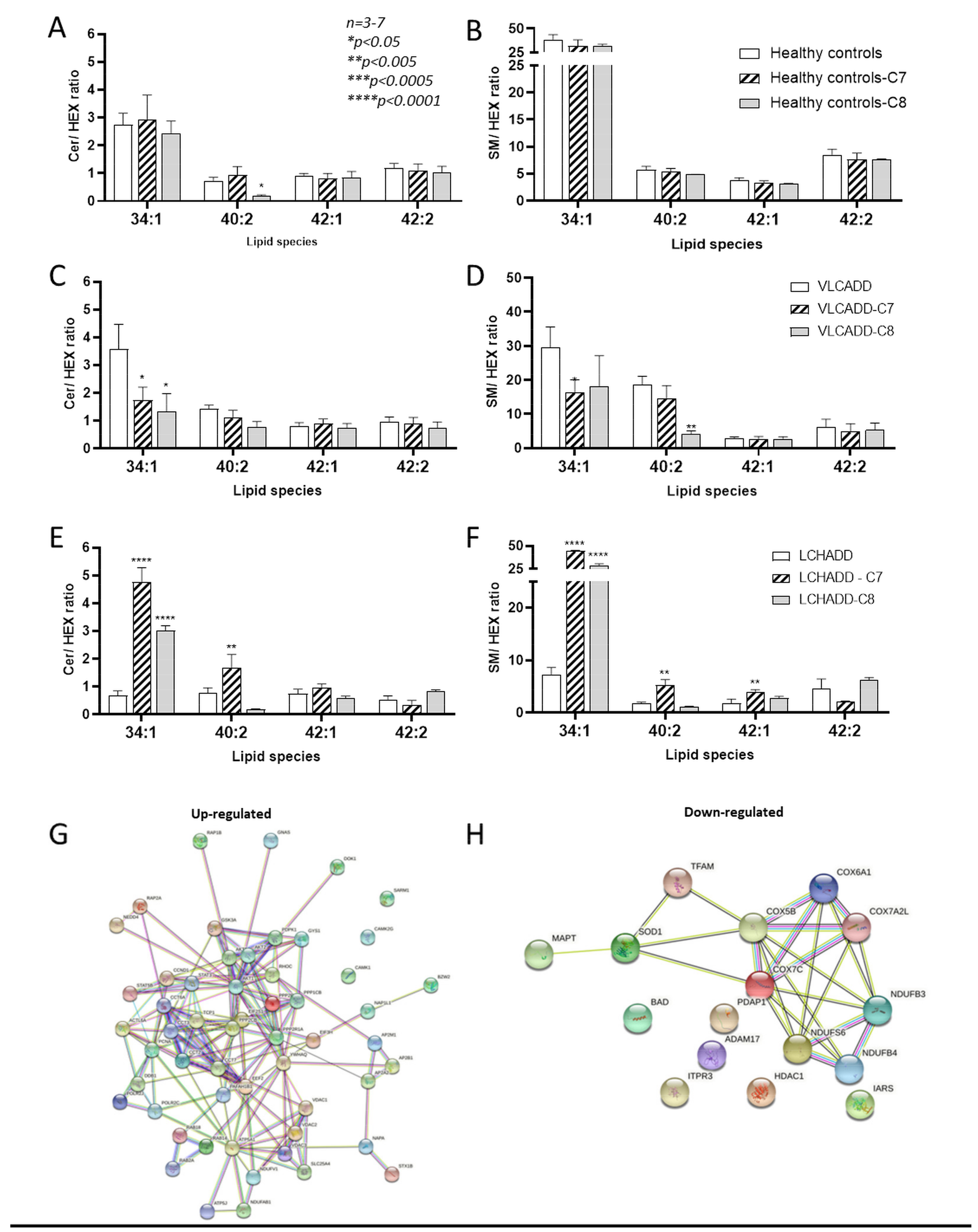
2.4. Effect of Mc-FAs on Sphingolipid Metabolic Flow
3. Discussion
Conclusions
4. Materials and Methods
4.1. Cell Culture
4.2. Lipidomic Analysis
4.3. Proteomic Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merritt, J.L., 2nd; Norris, M.; Kanungo, S. Fatty acid oxidation disorders. Ann. Transl Med. 2018, 6, 473. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.; Jackson, S.; Taroni, F.; Swift, P.; Turnbull, D.M. Characterisation of carnitine palmitoyltransferases in patients with a carnitine palmitoyltransferase deficiency: Implications for diagnosis and therapy. J. Neurol. Neurosurg. Psychiatry 1997, 62, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, G.L.; Van Hove, J.; Freedenberg, D.; Strauss, A.; Longo, N.; Burton, B.; Garganta, C.; Ficicioglu, C.; Cederbaum, S.; Harding, C.; et al. A delphi clinical practice protocol for the management of very long chain acyl-coa dehydrogenase deficiency. Mol. Genet. Metab. 2009, 96, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Spiekerkoetter, U.; Lindner, M.; Santer, R.; Grotzke, M.; Baumgartner, M.R.; Boehles, H.; Das, A.; Haase, C.; Hennermann, J.B.; Karall, D.; et al. Treatment recommendations in long-chain fatty acid oxidation defects: Consensus from a workshop. J. Inherit. Metab. Dis. 2009, 32, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Zand, D.; Doan, J.; Yi, S.; Wang, J.; Ma, L.; Akinshola, E.; Chakder, S.; Meyer, J.; Pacanowski, M.; Johnson, L.L.; et al. Regulatory news: Dojolvi (triheptanoin) as a source of calories and fatty acids in long-chain fatty acid oxidation disorders: Fda approval summary. J. Inherit. Metab. Dis. 2021, 44, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Lemarie, F.; Beauchamp, E.; Legrand, P.; Rioux, V. Revisiting the metabolism and physiological functions of caprylic acid (c8:0) with special focus on ghrelin octanoylation. Biochimie 2016, 120, 40–48. [Google Scholar] [CrossRef]
- Bach, A.C.; Babayan, V.K. Medium-chain triglycerides: An update. Am. J. Clin. Nutr. 1982, 36, 950–962. [Google Scholar] [CrossRef]
- Deng, S.; Zhang, G.F.; Kasumov, T.; Roe, C.R.; Brunengraber, H. Interrelations between c4 ketogenesis, c5 ketogenesis, and anaplerosis in the perfused rat liver. J. Biol. Chem. 2009, 284, 27799–27807. [Google Scholar] [CrossRef] [Green Version]
- Kinman, R.P.; Kasumov, T.; Jobbins, K.A.; Thomas, K.R.; Adams, J.E.; Brunengraber, L.N.; Kutz, G.; Brewer, W.U.; Roe, C.R.; Brunengraber, H. Parenteral and enteral metabolism of anaplerotic triheptanoin in normal rats. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E860–E866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin-Valencia, I.; Good, L.B.; Ma, Q.; Malloy, C.R.; Pascual, J.M. Heptanoate as a neural fuel: Energetic and neurotransmitter precursors in normal and glucose transporter i-deficient (g1d) brain. J. Cereb. Blood Flow Metab. 2013, 33, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Tucci, S.; Behringer, S.; Spiekerkoetter, U. De novo fatty acid biosynthesis and elongation in very long-chain acyl-coa dehydrogenase- (vlcad) deficient mice supplemented with odd or even medium-chain fatty acids. FEBS J. 2015, 282, 4242–4253. [Google Scholar] [CrossRef] [Green Version]
- Tucci, S.; Floegel, U.; Beermann, F.; Behringer, S.; Spiekerkoetter, U. Triheptanoin: Long-term effects in the very long-chain acyl-coa dehydrogenase (vlcad−/−)-deficient mouse. J. Lipid Res. 2016, 58, 196–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Mohsen, A.W.; Mihalik, S.J.; Goetzman, E.S.; Vockley, J. Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. J. Biol Chem 2010, 285, 29834–29841. [Google Scholar] [CrossRef] [Green Version]
- Ahmadpour, S.T.; Maheo, K.; Servais, S.; Brisson, L.; Dumas, J.F. Cardiolipin, the mitochondrial signature lipid: Implication in cancer. Int. J. Mol. Sci. 2020, 21, 8031. [Google Scholar] [CrossRef]
- Taylor, W.A.; Mejia, E.M.; Mitchell, R.W.; Choy, P.C.; Sparagna, G.C.; Hatch, G.M. Human trifunctional protein alpha links cardiolipin remodeling to beta-oxidation. PLoS ONE 2012, 7, e48628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Bharathi, S.S.; Rardin, M.J.; Uppala, R.; Verdin, E.; Gibson, B.W.; Goetzman, E.S. Sirt3 and sirt5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-coa dehydrogenase. PLoS ONE 2015, 10, e0122297. [Google Scholar] [CrossRef] [Green Version]
- Hagenbuchner, J.; Scholl-Buergi, S.; Karall, D.; Ausserlechner, M.J. Very long-/ and long chain-3-hydroxy acyl coa dehydrogenase deficiency correlates with deregulation of the mitochondrial fusion/fission machinery. Sci. Rep. 2018, 8, 3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimo, S.; Zura-Miller, G.; Fezelinia, H.; Spruce, L.A.; Zakopoulos, I.; Mohsen, A.W.; Vockley, J.; Ischiropoulos, H. Mitochondrial morphology, bioenergetics and proteomic responses in fatty acid oxidation disorders. Redox Biol. 2021, 41, 101923. [Google Scholar] [CrossRef]
- Alatibi, K.I.; Hagenbuchner, J.; Wehbe, Z.; Karall, D.; Ausserlechner, M.J.; Vockley, J.; Spiekerkoetter, U.; Grunert, S.C.; Tucci, S. Different lipid signature in fibroblasts of long-chain fatty acid oxidation disorders. Cells 2021, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Checa, A.; Khademi, M.; Sar, D.G.; Haeggstrom, J.Z.; Lundberg, J.O.; Piehl, F.; Olsson, T.; Wheelock, C.E. Hexosylceramides as intrathecal markers of worsening disability in multiple sclerosis. Mult. Scler. 2015, 21, 1271–1279. [Google Scholar] [CrossRef]
- Mielke, M.M.; Maetzler, W.; Haughey, N.J.; Bandaru, V.V.; Savica, R.; Deuschle, C.; Gasser, T.; Hauser, A.K.; Graber-Sultan, S.; Schleicher, E.; et al. Plasma ceramide and glucosylceramide metabolism is altered in sporadic parkinson’s disease and associated with cognitive impairment: A pilot study. PLoS ONE 2013, 8, e73094. [Google Scholar] [CrossRef] [Green Version]
- Pujol-Lereis, L.M. Alteration of sphingolipids in biofluids: Implications for neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 3564. [Google Scholar] [CrossRef] [Green Version]
- Vidaurre, O.G.; Haines, J.D.; Katz Sand, I.; Adula, K.P.; Huynh, J.L.; McGraw, C.A.; Zhang, F.; Varghese, M.; Sotirchos, E.; Bhargava, P.; et al. Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain 2014, 137, 2271–2286. [Google Scholar] [CrossRef]
- Wan, S.; Kuipers, F.; Havinga, R.; Ando, H.; Vance, D.E.; Jacobs, R.L.; van der Veen, J.N. Impaired hepatic phosphatidylcholine synthesis leads to cholestasis in mice challenged with a high-fat diet. Hepatol. Commun. 2019, 3, 262–276. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom Rev. 2015, 34, 474–490. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.; Mann, M. Bioinformatics analysis of mass spectrometry-based proteomics data sets. FEBS Lett. 2009, 583, 1703–1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreutzberger, A.J.B.; Ji, M.; Aaron, J.; Mihaljevic, L.; Urban, S. Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science 2019, 363, eaao0076. [Google Scholar] [CrossRef]
- Sezgin, E.; Gutmann, T.; Buhl, T.; Dirkx, R.; Grzybek, M.; Coskun, U.; Solimena, M.; Simons, K.; Levental, I.; Schwille, P. Adaptive lipid packing and bioactivity in membrane domains. PLoS ONE 2015, 10, e0123930. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Jatooratthawichot, P.; Talabnin, C.; Ngiwsara, L.; Rustam, Y.H.; Svasti, J.; Reid, G.E.; Ketudat Cairns, J.R. Effect of expression of human glucosylceramidase 2 isoforms on lipid profiles in cos-7 cells. Metabolites 2020, 10, 488. [Google Scholar] [CrossRef]
- Guffon, N.; Mochel, F.; Schiff, M.; De Lonlay, P.; Douillard, C.; Vianey-Saban, C. Clinical outcomes in a series of 18 patients with long chain fatty acids oxidation disorders treated with triheptanoin for a median duration of 22 months. Mol. Genet. Metab. 2021, 132, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Vockley, J.; Burton, B.; Berry, G.; Longo, N.; Phillips, J.; Sanchez-Valle, A.; Chapman, K.; Tanpaiboon, P.; Grunewald, S.; Murphy, E.; et al. Effects of triheptanoin (ux007) in patients with long-chain fatty acid oxidation disorders: Results from an open-label, long-term extension study. J. Inherit. Metab. Dis. 2020, 44, 253–263. [Google Scholar] [CrossRef]
- Vockley, J.; Burton, B.; Berry, G.T.; Longo, N.; Phillips, J.; Sanchez-Valle, A.; Tanpaiboon, P.; Grunewald, S.; Murphy, E.; Humphrey, R.; et al. Ux007 for the treatment of long chain-fatty acid oxidation disorders: Safety and efficacy in children and adults following 24weeks of treatment. Mol. Genet. Metab. 2017, 120, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Zoggeler, T.; Stock, K.; Jorg-Streller, M.; Spenger, J.; Konstantopoulou, V.; Hufgard-Leitner, M.; Scholl-Burgi, S.; Karall, D. Long-term experience with triheptanoin in 12 austrian patients with long-chain fatty acid oxidation disorders. Orphanet. J. Rare Dis. 2021, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. Mapt mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Invest. 2019, 99, 912–928. [Google Scholar] [CrossRef]
- Cluskey, S.; Ramsden, D.B. Mechanisms of neurodegeneration in amyotrophic lateral sclerosis. Mol. Pathol. 2001, 54, 386–392. [Google Scholar] [PubMed]
- Trist, B.G.; Hare, D.J.; Double, K.L. A proposed mechanism for neurodegeneration in movement disorders characterized by metal dyshomeostasis and oxidative stress. Cell Chem. Biol. 2018, 25, 807–816. [Google Scholar] [CrossRef]
- Kang, I.; Chu, C.T.; Kaufman, B.A. The mitochondrial transcription factor tfam in neurodegeneration: Emerging evidence and mechanisms. FEBS Lett. 2018, 592, 793–811. [Google Scholar] [CrossRef] [Green Version]
- Ban, T.; Ishihara, T.; Kohno, H.; Saita, S.; Ichimura, A.; Maenaka, K.; Oka, T.; Mihara, K.; Ishihara, N. Molecular basis of selective mitochondrial fusion by heterotypic action between opa1 and cardiolipin. Nat. Cell Biol. 2017, 19, 856–863. [Google Scholar] [CrossRef]
- Kameoka, S.; Adachi, Y.; Okamoto, K.; Iijima, M.; Sesaki, H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018, 28, 67–76. [Google Scholar] [CrossRef]
- Musatov, A.; Sedlak, E. Role of cardiolipin in stability of integral membrane proteins. Biochimie 2017, 142, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Claypool, S.M.; Koehler, C.M. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 2012, 37, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudek, J. Role of cardiolipin in mitochondrial signaling pathways. Front. Cell Dev. Biol. 2017, 5, 90. [Google Scholar] [CrossRef] [Green Version]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Cardiolipin and mitochondrial function in health and disease. Antioxid Redox Signal. 2014, 20, 1925–1953. [Google Scholar] [CrossRef]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of cardiolipin in mitochondrial function and dynamics in health and disease: Molecular and pharmacological aspects. Cells 2019, 8, 728. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Pulakat, L.; Whaley-Connell, A.; Sowers, J.R. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J. Mol. Med. 2010, 88, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- Wasmus, C.; Dudek, J. Metabolic alterations caused by defective cardiolipin remodeling in inherited cardiomyopathies. Life 2020, 10, 277. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Casares, D.; Escriba, P.V.; Rossello, C.A. Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piomelli, D.; Astarita, G.; Rapaka, R. A neuroscientist’s guide to lipidomics. Nat. Rev. Neurosci. 2007, 8, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Herrera, F.; Taoro-Gonzalez, L.; Valdes-Baizabal, C.; Diaz, M.; Marin, R. Lipid and lipid raft alteration in aging and neurodegenerative diseases: A window for the development of new biomarkers. Int. J. Mol. Sci. 2019, 20, 3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernert, J.T., Jr.; Ullman, M.D. Biosynthesis of sphingomyelin from erythro-ceramides and phosphatidylcholine by a microsomal cholinephosphotransferase. Biochim. Biophys. Acta 1981, 666, 99–109. [Google Scholar] [CrossRef]
- Ullman, M.D.; Radin, N.S. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J. Biol. Chem. 1974, 249, 1506–1512. [Google Scholar] [CrossRef]
- Villani, M.; Subathra, M.; Im, Y.B.; Choi, Y.; Signorelli, P.; Del Poeta, M.; Luberto, C. Sphingomyelin synthases regulate production of diacylglycerol at the golgi. Biochem. J. 2008, 414, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Voelker, D.R.; Kennedy, E.P. Cellular and enzymic synthesis of sphingomyelin. Biochemistry 1982, 21, 2753–2759. [Google Scholar] [CrossRef]
- Grunert, S.C.; Eckenweiler, M.; Haas, D.; Lindner, M.; Tsiakas, K.; Santer, R.; Tucci, S.; Spiekerkoetter, U. The spectrum of peripheral neuropathy in disorders of the mitochondrial trifunctional protein. J. Inherit. Metab. Dis. 2021, 44, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.A.; Majumder, S.; Zhu, H.; Lee, Y.T.; Kono, M.; Li, C.; Khanna, C.; Blain, H.; Schwartz, R.; Huso, V.L.; et al. The ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. Elife 2019, 8, e51067. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Bogdanov, M.; Mileykovskaya, E.; Dowhan, W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: Implications for lipid-linked disorders. Subcell. Biochem. 2008, 49, 197–239. [Google Scholar]
- Novack, G.V.; Galeano, P.; Castano, E.M.; Morelli, L. Mitochondrial supercomplexes: Physiological organization and dysregulation in age-related neurodegenerative disorders. Front. Endocrinol. 2020, 11, 600. [Google Scholar] [CrossRef]
- Solsona-Vilarrasa, E.; Fucho, R.; Torres, S.; Nunez, S.; Nuno-Lambarri, N.; Enrich, C.; Garcia-Ruiz, C.; Fernandez-Checa, J.C. Cholesterol enrichment in liver mitochondria impairs oxidative phosphorylation and disrupts the assembly of respiratory supercomplexes. Redox Biol. 2019, 24, 101214. [Google Scholar] [CrossRef]
- Hesse, J.; Braun, C.; Behringer, S.; Matysiak, U.; Spiekerkoetter, U.; Tucci, S. The diagnostic challenge in very-long chain acyl-coa dehydrogenase deficiency (vlcadd). J. Inherit. Metab. Dis. 2018, 41, 1169–1178. [Google Scholar] [CrossRef]
- Sampaio, J.L.; Gerl, M.J.; Klose, C.; Ejsing, C.S.; Beug, H.; Simons, K.; Shevchenko, A. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA 2011, 108, 1903–1907. [Google Scholar] [CrossRef] [Green Version]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar] [CrossRef] [Green Version]
- Surma, M.A.; Herzog, R.; Vasilj, A.; Klose, C.; Christinat, N.; Morin-Rivron, D.; Simons, K.; Masoodi, M.; Sampaio, J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. EJLST 2015, 117, 1540–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, R.; Schuhmann, K.; Schwudke, D.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. Lipidxplorer: A software for consensual cross-platform lipidomics. PLoS ONE 2012, 7, e29851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, R.; Schwudke, D.; Schuhmann, K.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 2011, 12, R8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alatibi, K.I.; Wehbe, Z.; Spiekerkoetter, U.; Tucci, S. Sex-specific perturbation of complex lipids in response to medium-chain fatty acids in very long-chain acyl-coa dehydrogenase deficiency. FEBS J. 2020, 287, 3511–3525. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.; Schafer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Johnstone, R.; Mohammed, A.K.; Hamon, C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by ms/ms. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef] [PubMed]

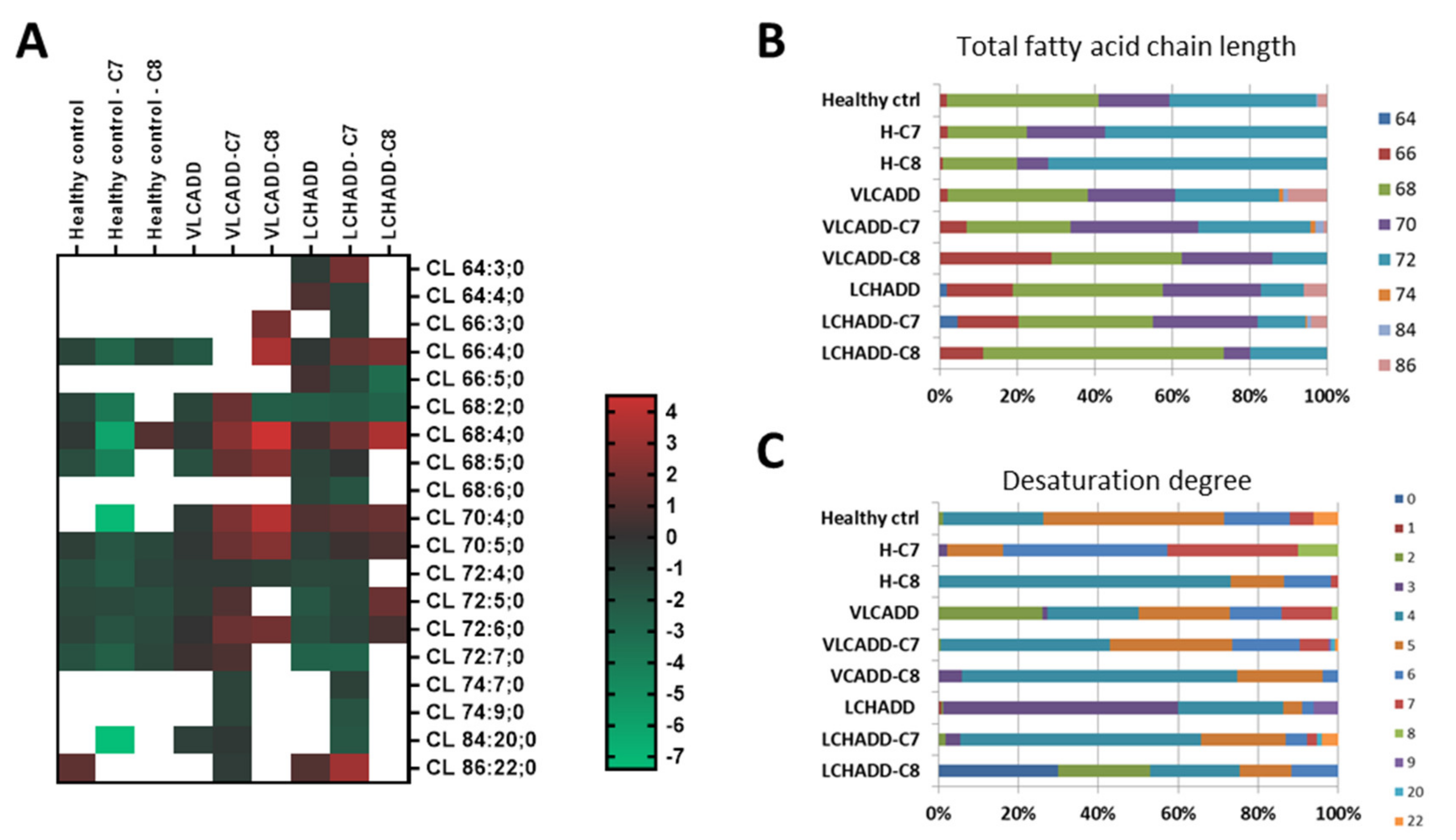
| PC/PE Ratio | |||
|---|---|---|---|
| Control Conditions a | C7 | C8 | |
| Healthy controls | 3.78 | 4.07 | 3.92 |
| VLCADD | 3.85 | 4.83 * | 4.23 |
| LCHADD | 4.33 | 3.96 | 2.9 ** |
| Comparison | Number of Proteins Altered | Up-Regulated | Down-Regulated |
|---|---|---|---|
| Healthy control vs LCHADD | 338 | 235 | 103 |
| Healthy control vs VLCADD | 104 | 48 | 56 |
| Healthy control-C7 vs LCHADD-C7 | 922 | 406 | 516 |
| Healthy control-C7 vs VLCADD-C8 | 232 | 146 | 86 |
| Healthy control-C8 vs LCHADD-C8 | 231 | 115 | 116 |
| Healthy control-C8 vs VLCADD-C8 | 312 | 90 | 122 |
| Healthy control vs Healthy control-C7 | 36 | 5 | 31 |
| Healthy control vs Healthy control-C8 | 7 | 7 | |
| LCHADD vs LCHADD-C7 | 81 | 27 | 54 |
| LCHADD vs LCHADD-C8 | 25 | 16 | 9 |
| VLCADD vs VLCADD-C7 | 2 | 2 | |
| VLCADD vs VLCADD-C8 | 9 | 5 | 4 |
| Disease | Origin | Sex | Allele 1 | Allele 2 |
|---|---|---|---|---|
| Healthy control | Coriell Institute (GM04501) | m | WT | WT |
| Healthy control | Coriell Institute (GM04505) | f | WT | WT |
| Healthy control | Coriell Institute (GM07492) | m | WT | WT |
| Healthy control | Coriell Institute (GM08399) | f | WT | WT |
| Healthy control | Coriell Institute (GM08400) | f | WT | WT |
| Healthy control | Coriell Institute (GM23964) | m | WT | WT |
| Healthy control | Coriell Institute (GM23976) | m | WT | WT |
| VLCAD | Coriell Institute (GM06127) | m | c.925G > a | c.925G > A |
| VLCAD | Coriell Institute (GM09093) | f | c.515T > C | c.637G > A |
| VLCAD | Coriell Institute (GM17475) | m | c.364A > G | c.364A > G |
| VLCAD | Prof. Dr. J. Vockley | f | c.1619T > C | c.1707_1715dupAGACGGGGC |
| VLCAD | Prof. Dr. J. Vockley | m | c.520G > A | c.1825G > A |
| VLCAD | Prof. Dr. J. Vockley | f | c.14T > C | c.1182 + 2dupT |
| LCHAD | Prof. Dr. D. Karall | m | c.1528G < C | c.1528G < C |
| LCHAD | Prof. Dr. D. Karall | f | c.1528G < C | c.1528G < C |
| LCHAD | Prof. Dr. D. Karall | m | c.1528G < C | c.1528G < C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alatibi, K.I.; Tholen, S.; Wehbe, Z.; Hagenbuchner, J.; Karall, D.; Ausserlechner, M.J.; Schilling, O.; Grünert, S.C.; Vockley, J.; Tucci, S. Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders. Int. J. Mol. Sci. 2021, 22, 10556. https://doi.org/10.3390/ijms221910556
Alatibi KI, Tholen S, Wehbe Z, Hagenbuchner J, Karall D, Ausserlechner MJ, Schilling O, Grünert SC, Vockley J, Tucci S. Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders. International Journal of Molecular Sciences. 2021; 22(19):10556. https://doi.org/10.3390/ijms221910556
Chicago/Turabian StyleAlatibi, Khaled I., Stefan Tholen, Zeinab Wehbe, Judith Hagenbuchner, Daniela Karall, Michael J. Ausserlechner, Oliver Schilling, Sarah C. Grünert, Jerry Vockley, and Sara Tucci. 2021. "Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders" International Journal of Molecular Sciences 22, no. 19: 10556. https://doi.org/10.3390/ijms221910556
APA StyleAlatibi, K. I., Tholen, S., Wehbe, Z., Hagenbuchner, J., Karall, D., Ausserlechner, M. J., Schilling, O., Grünert, S. C., Vockley, J., & Tucci, S. (2021). Lipidomic and Proteomic Alterations Induced by Even and Odd Medium-Chain Fatty Acids on Fibroblasts of Long-Chain Fatty Acid Oxidation Disorders. International Journal of Molecular Sciences, 22(19), 10556. https://doi.org/10.3390/ijms221910556







