Post-Embryonic Lateral Organ Development and Adaxial—Abaxial Polarity Are Regulated by the Combined Effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis Shoots
Abstract
1. Introduction
2. Results
2.1. esr1 Mutations Enhanced Defects in Rosette Leaf Development and Adaxial—Abaxial Polarity in wus Background
2.2. wus-101 esr1-1 Phenocopied wus-101 rev-5
2.3. esr1-1 and esr2-2 Antagonistically Regulate Rosette Leaf Development in bum1-3 in the Later Vegetative Phase
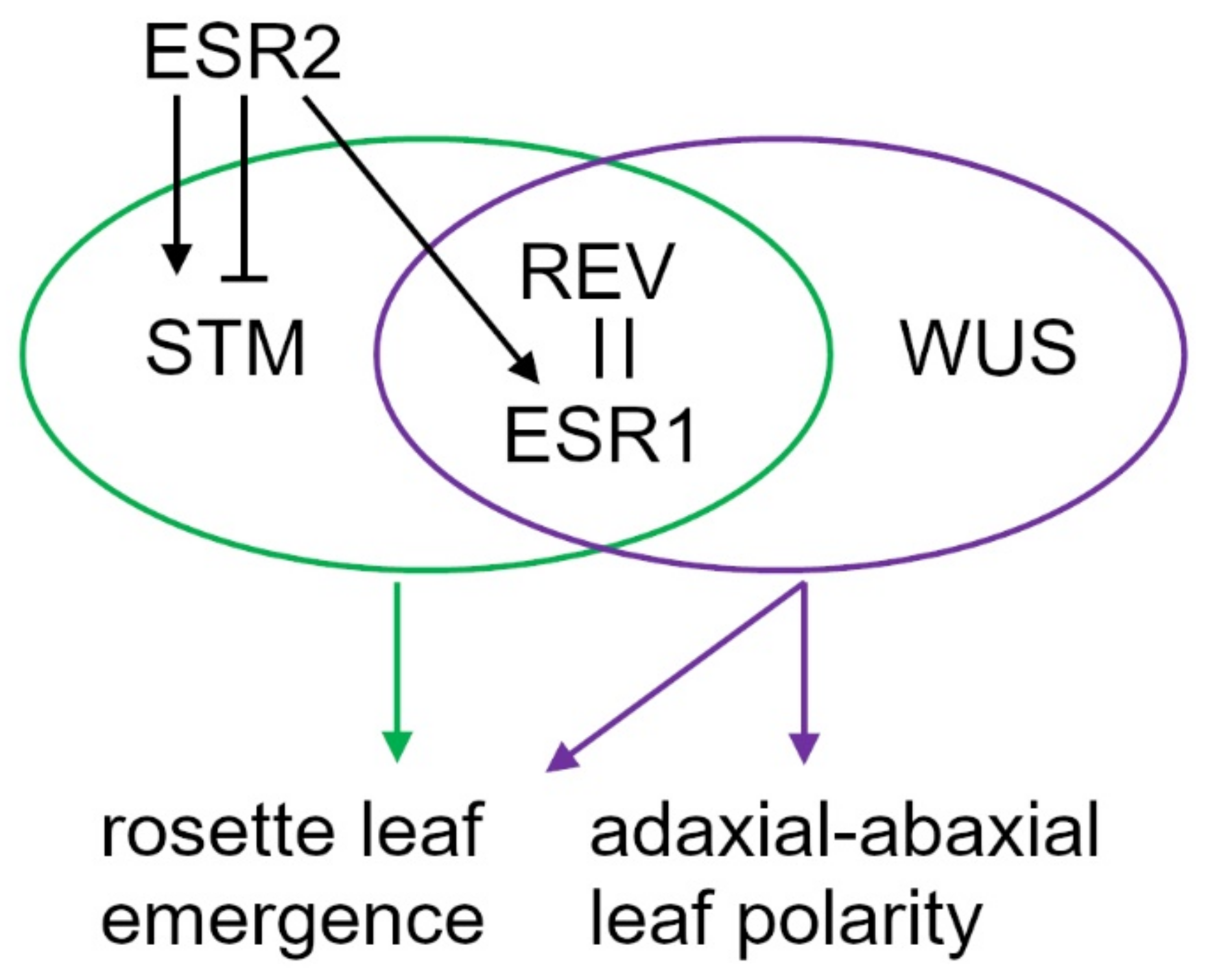
2.4. Distinct Expression Pattern of ESR Genes Defines their Unique Roles
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Construction of Transgene and Transformation
4.3. Semi-Quantitative RT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.X.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Clark, S.E. A WUSCHEL-independent stem cell specification pathway is repressed by PHB, PHV and CNA in arabidopsis. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef]
- Huang, W.; Pitorre, D.; Poretska, O.; Marizzi, C.; Winter, N.; Poppenberger, B.; Sieberer, T. Altered meristem programSuppresses ectopic stem cell niche formation in the shoot apical meristem in a largely cytokinin-independent manner. Plant Physiol. 2015, 167, 1471–1486. [Google Scholar] [CrossRef]
- Jung, J.H.; Park, C.M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007, 225, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Grigg, S.P.; Xie, M.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Otsuga, D.; DeGuzman, B.; Prigge, M.J.; Drews, G.N.; Clark, S.E. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001, 25, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial Patterning of Arabidopsis Shoots by Class III HD-ZIP and KANADI Genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef]
- Banno, H.; Ikeda, Y.; Niu, Q.W.; Chua, N.H. Overexpression of Arabidopsis ESR1 induces initiation of Shoot Regeneration. Plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef]
- Kirch, T.; Simon, R.; Grünewald, M.; Werr, W. The Dornröschen/enhancer of shoot regeneration1: Gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 2003, 15, 694–705. [Google Scholar] [CrossRef]
- Ikeda, Y.; Banno, H.; Niu, Q.-W.; Howell, S.H.; Chua, N.-H. The enhancer of shoot regeneration 2 gene in Arabidopsis regulates cup-shaped cotyledon 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006, 47, 1443–1456. [Google Scholar] [CrossRef]
- Ward, J.M.; Smith, A.M.; Shah, P.K.; Galanti, S.E.; Yi, H.; Demianski, A.J.; Van Der Graaff, E.; Keller, B.; Neff, M.M. A new role for the Arabidopis AP2 transcription factor, leafy petiole, in gibberellin-induced germination is revealed by the misexpression of a homologous gene SOB2/DRN-LIKE. Plant Cell 2006, 18, 29–39. [Google Scholar] [CrossRef][Green Version]
- Marsch-Martinez, N.; Greco, R.; Becker, J.D.; Dixit, S.; Bergervoet, J.H.W.; Karaba, A.; De Folter, S.; Pereira, A. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 2006, 62, 825–843. [Google Scholar] [CrossRef] [PubMed]
- Capua, Y.; Eshed, Y. Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. Proc. Natl. Acad. Sci. USA 2017, 114, 3246–3251. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Cole, M.; Flier, A.; Grewe, B.; Werr, W. The AP2 transcription factors DORNRÖSCHEN and Dornröschen-like redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 2007, 134, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Durán-Medina, Y.; Serwatowska, J.; Reyes-Olalde, J.I.; De Folter, S.; Marsch-Martínez, N. The AP2/ERF transcription factor DRNL modulates gynoecium development and affects its response to Cytokinin. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Nag, A.; Yang, Y.; Jack, T. DORNRÖSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Mol. Biol. 2007, 65, 219–232. [Google Scholar] [CrossRef]
- Matsuo, N.; Makino, M.; Banno, H. Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 2011, 181, 39–46. [Google Scholar] [CrossRef]
- Matsuo, N.; Mase, H.; Makino, M.; Takahashi, H.; Banno, H. Identification of enhancer of shoot regeneration 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 2009, 26, 385–393. [Google Scholar] [CrossRef]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J.; Wenkel, S.; Chandler, J.W.; Werr, W.; Jiao, Y. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 2018, 145, dev158352. [Google Scholar] [CrossRef] [PubMed]
- Kubalová, I.; Zalabák, D.; Mičúchová, A.; Ikeda, Y. Mutations in tetrapyrrole biosynthesis pathway uncouple nuclear WUSCHEL expression from de novo shoot development in Arabidopsis. Plant Cell Tissue Organ Cult. 2019, 139, 395–401. [Google Scholar] [CrossRef]
- Magnani, E.; Barton, M.K. A Per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 2011, 23, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Niihama, M.; Smith, H.M.S.; Tasaka, M.; Aida, M. Gorgon, a novel missense mutation in the shoot meristemless gene, impairs shoot meristem homeostasis in arabidopsis. Plant Cell Physiol. 2010, 51, 621–634. [Google Scholar] [CrossRef]
- Campisi, L.; Yang, Y.; Yi, Y.; Heilig, E.; Herman, B.; Cassista, A.J.; Allen, D.W.; Xiang, H.; Jack, T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999, 17, 699–707. [Google Scholar] [CrossRef]
- Glowa, D.; Comelli, P.; Chandler, J.W.; Werr, W. Clonal sector analysis and cell ablation confirm a function for DORNROESCHEN-LIKE in founder cells and the vasculature in Arabidopsis. Planta 2021, 253, 1–16. [Google Scholar] [CrossRef]
- Caggiano, M.P.; Yu, X.; Bhatia, N.; Larsson, A.; Ram, H.; Ohno, C.K.; Sappl, P.; Meyerowitz, E.M.; Jönsson, H.; Heisler, M.G. Cell type boundaries organize plant development. Elife 2017, 6, 1–32. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of pluripotency pathways regulates stem cell maintenance in the arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xu, L.; Xu, B.; Yang, L.; Ling, Q.; Wang, H.; Huang, H. Genetic interactions between leaf polarity-controlling genes and ASYMMETRIC LEAVES1 and 2 in Arabidopsis leaf patterning. Plant Cell Physiol. 2007, 48, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, Y.; Dong, A.; Sun, Y.; Pi, L.; Xu, Y.; Huang, H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003, 130, 4097–4107. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Sun, Y.; Xu, L.; Xu, Y.; Huang, H. ERECTA is required for protection against heat-stress in the AS1/AS2 pathway to regulate adaxial-abaxial leaf polarity in Arabidopsis. Planta 2004, 219, 270–276. [Google Scholar] [CrossRef]
- Xu, M.; Du, Q.; Tian, C.; Wang, Y.; Jiao, Y. Stochastic gene expression drives mesophyll protoplast regeneration. Sci. Adv. 2021, 7, eabg8466. [Google Scholar] [CrossRef]
- Xiang, D.; Yang, H.; Venglat, P.; Cao, Y.; Wen, R.; Ren, M.; Stone, S.; Wang, E.; Wang, H.; Xiao, W.; et al. POPCORN functions in the auxin pathway to regulate embryonic body plan and meristem organization in Arabidopsis. Plant Cell 2011, 23, 4348–4367. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jurgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Turchi, L.; Carabelli, M.; Ruzza, V.; Possenti, M.; Sassi, M.; Peñalosa, A.; Sessa, G.; Salvi, S.; Forte, V.; Morelli, G.; et al. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013, 140, 2118–2129. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]

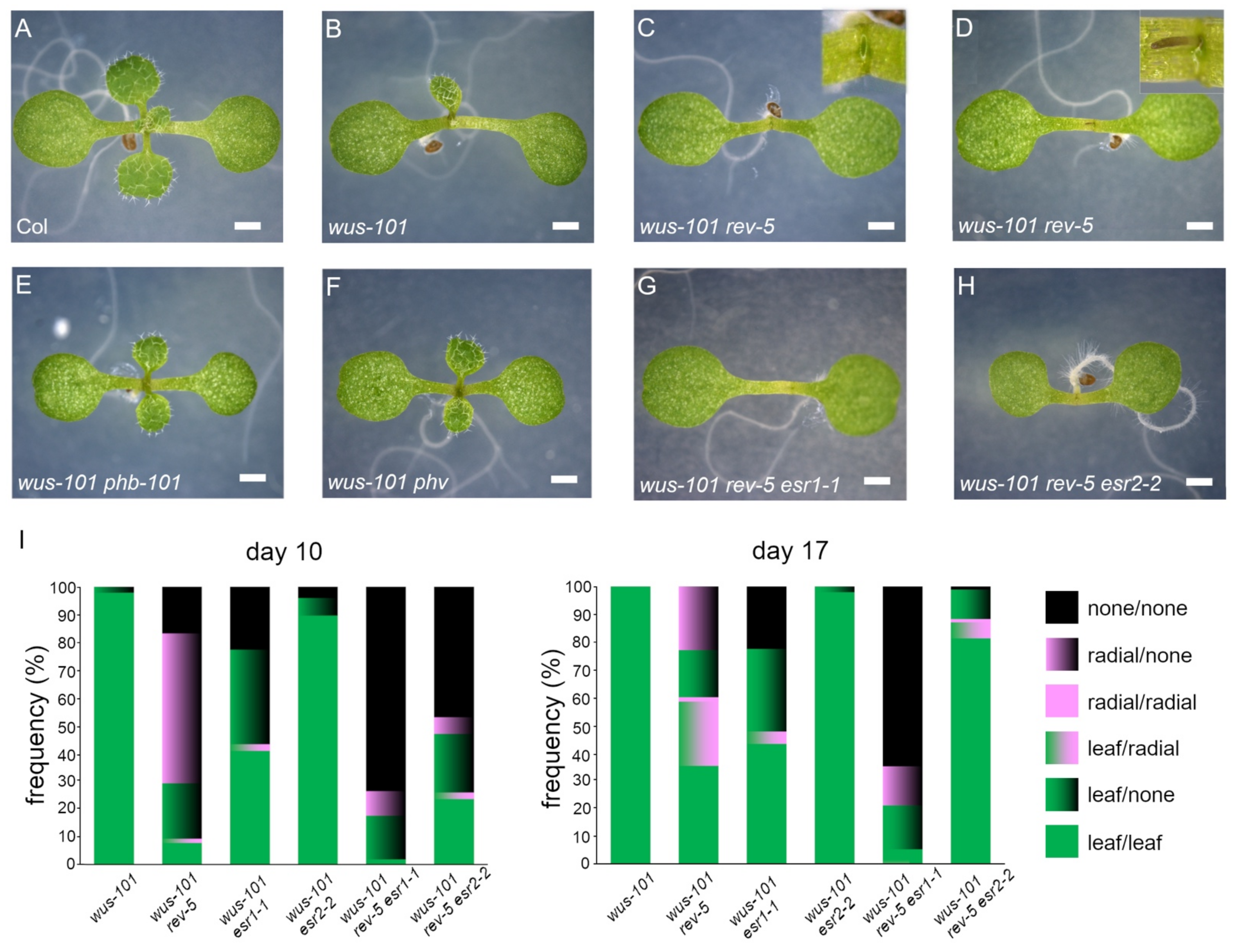


| wus b | Radial Structure d | Unrecognizable True Leaves | mp-Like | pin-Like Shoot | |
|---|---|---|---|---|---|
| wus-1 (n = 160) | 100 | 0 | 0 | 0 | 0 |
| wus-1;esr1-1 (n = 54) | 68.5 c | 14.8 | 14.8 e | 1.9 | 0 |
| wus-101 (n = 196) | 100 | 0 | 0 | 0 | 0 |
| wus-101;esr1-1 (n = 88) | 35.2 c | 9.1 | 55.7 e | 0 | 0 |
| wus-101;esr1-2 (n = 38) | 78.9 c | 21.1 | 0 | 0 | 0 |
| wus-101;esr2-2 (n = 55) | 97.9 | 0 | 2.1 | 0 | 0 |
| esr1-1;esr2-2e (n = 47) | 0 | 0 | 0 | 26.1 | 0 |
| wus-101;esr1-1;esr2-2 (n = 72) | 9.7 c | 5.6 | 34.7 f | 38.9 | 11.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, Y.; Králová, M.; Zalabák, D.; Kubalová, I.; Aida, M. Post-Embryonic Lateral Organ Development and Adaxial—Abaxial Polarity Are Regulated by the Combined Effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis Shoots. Int. J. Mol. Sci. 2021, 22, 10621. https://doi.org/10.3390/ijms221910621
Ikeda Y, Králová M, Zalabák D, Kubalová I, Aida M. Post-Embryonic Lateral Organ Development and Adaxial—Abaxial Polarity Are Regulated by the Combined Effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis Shoots. International Journal of Molecular Sciences. 2021; 22(19):10621. https://doi.org/10.3390/ijms221910621
Chicago/Turabian StyleIkeda, Yoshihisa, Michaela Králová, David Zalabák, Ivona Kubalová, and Mitsuhiro Aida. 2021. "Post-Embryonic Lateral Organ Development and Adaxial—Abaxial Polarity Are Regulated by the Combined Effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis Shoots" International Journal of Molecular Sciences 22, no. 19: 10621. https://doi.org/10.3390/ijms221910621
APA StyleIkeda, Y., Králová, M., Zalabák, D., Kubalová, I., & Aida, M. (2021). Post-Embryonic Lateral Organ Development and Adaxial—Abaxial Polarity Are Regulated by the Combined Effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis Shoots. International Journal of Molecular Sciences, 22(19), 10621. https://doi.org/10.3390/ijms221910621







