The FMN “140s Loop” of Cytochrome P450 Reductase Controls Electron Transfer to Cytochrome P450
Abstract
:1. Introduction
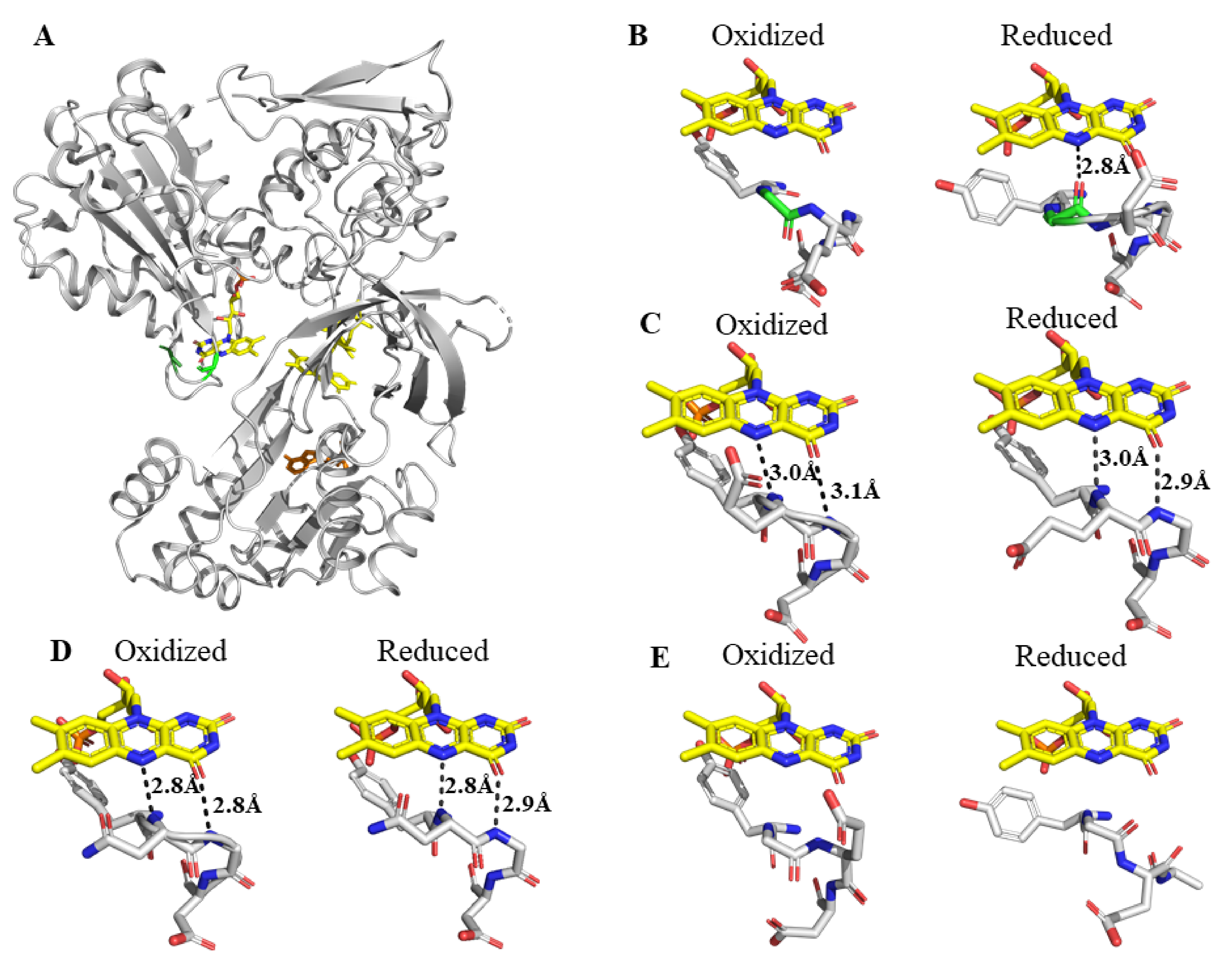
2. Results
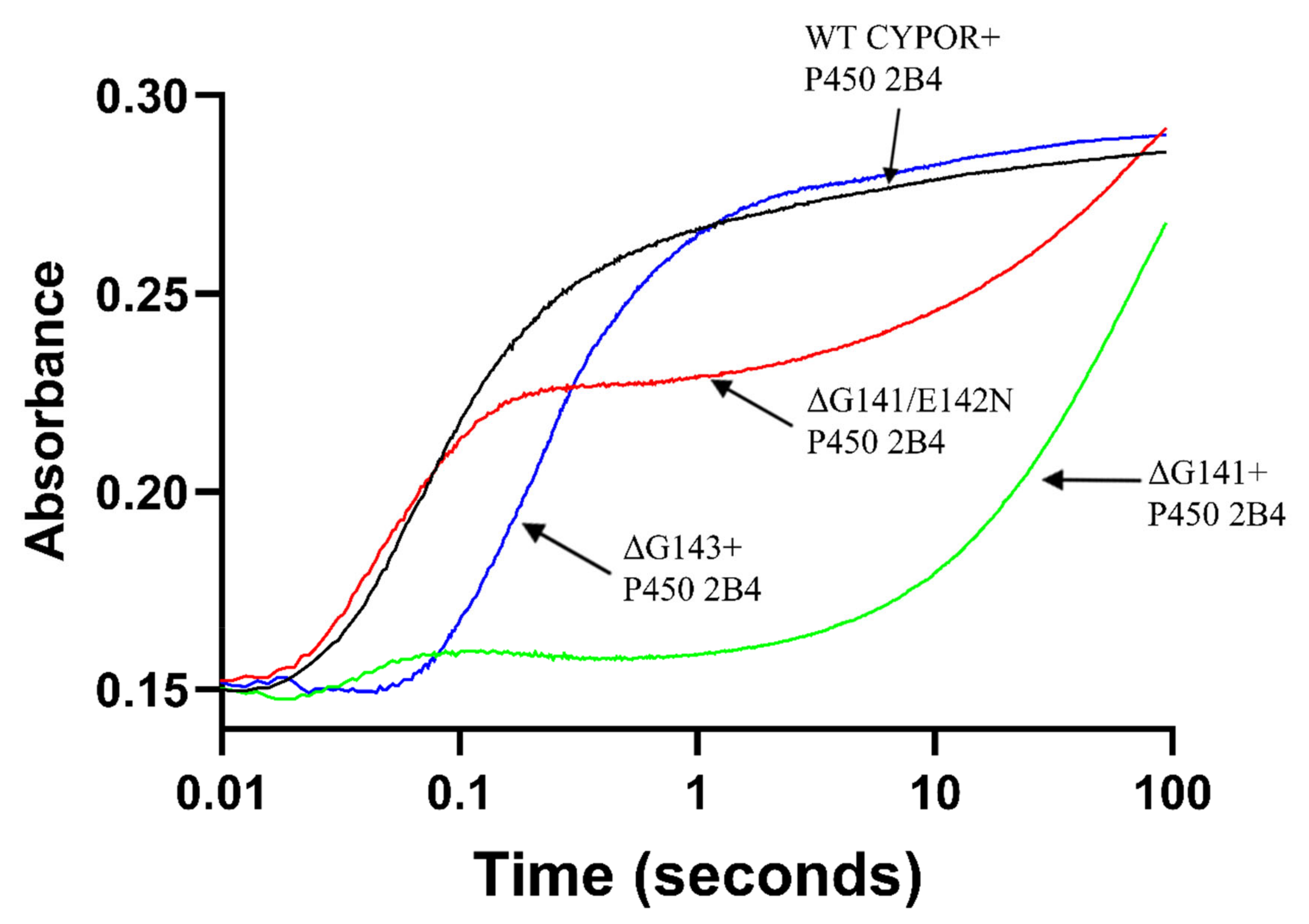
2.1. Comparison of the Kinetics of Reduction of Ferric Cytochrome P450 by Wild Type and Mutant Reductases
2.2. Effect of Cytochrome b5 on the Rate of Benzphetamine Metabolism by Cyt P450 2B4 under Steady-State Conditions at 30 °C in the Presence of Wild-Type and Mutant Reductases
3. Discussion
4. Materials and Methods
4.1. Construction of Site-Directed Mutants of the Full-Length CYPOR
4.2. Expression and Purification of the Full-Length Wild-Type (WT) and Glycine Deletion CYPOR Mutants
4.3. Measurement of Benzphetamine Metabolism by Cyt P450 2B4
4.4. Kinetics of the Reduction of Ferric Cyt P450 2B4 by CYPOR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Iyanagi, T.; Xia, C.; Kim, J.-J.P. NADPH-cytochrome P450 oxidoreductase: Prototypic member of the diflavin reductase family. Arch. Biochem. Biophys. 2012, 528, 72–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waskell, L.; Kim, J.-J.P. Electron Transfer Partners of Cytochrome P450. In Cytochrome P450; Springer International Publishing: Cham, Switzerland, 2015; pp. 33–68. [Google Scholar]
- Iyanagi, T. Molecular mechanism of metabolic NAD(P)H-dependent electron-transfer systems: The role of redox cofactors. Biochim. Biophys. Acta—Bioenerg. 2019, 1860, 233–258. [Google Scholar] [CrossRef] [PubMed]
- Miura, R. Versatility and specificity in flavoenzymes: Control mechanisms of flavin reactivity. Chem. Rec. 2001, 1, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Das, A.; Sibhatu, H.; Jamal, J.; Sligar, S.G.; Poulos, T.L. Exploring the electron transfer properties of neuronal nitric-oxide synthase by reversal of the FMN redox potential. J. Biol. Chem. 2008, 283, 34762–34772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guengerich, F.P. Human Cytochrome P450 Enzymes. In Cytochrome P450; Springer International Publishing: Cham, Switzerland, 2015; pp. 523–785. [Google Scholar]
- Ortiz de Montellano, P.R. Hydrocarbon Hydroxylation by Cytochrome P450 Enzymes. Chem. Rev. 2010, 110, 932–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šrejber, M.; Navrátilová, V.; Paloncýová, M.; Bazgier, V.; Berka, K.; Anzenbacher, P.; Otyepka, M. Membrane-attached mammalian cytochromes P450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners. J. Inorg. Biochem. 2018, 183, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Rwere, F.; Xia, C.; Im, S.; Haque, M.M.; Stuehr, D.J.; Waskell, L.; Kim, J.-J.P. Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone. J. Biol. Chem. 2016, 291, 14639–14661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley, S.C.; Ost, T.W.B.; Daff, S. The unusual redox properties of flavocytochrome P450 BM3 flavodoxin domain. Biochem. Biophys. Res. Commun. 2004, 325, 1418–1423. [Google Scholar] [CrossRef]
- Zhang, H.; Gruenke, L.; Arscott, D.; Shen, A.; Kasper, C.; Harris, D.L.; Glavanovich, M.; Johnson, R.; Waskell, L. Determination of the Rate of Reduction of Oxyferrous Cytochrome P450 2B4 by 5-Deazariboflavin Adenine Dinucleotide T491V Cytochrome P450 Reductase †. Biochemistry 2003, 42, 11594–11603. [Google Scholar] [CrossRef]
- Zhang, H.; Im, S.-C.; Waskell, L. Cytochrome b5 Increases the Rate of Product Formation by Cytochrome P450 2B4 and Competes with Cytochrome P450 Reductase for a Binding Site on Cytochrome P450 2B4. J. Biol. Chem. 2007, 282, 29766–29776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hamdane, D.; Im, S.-C.; Waskell, L. Cytochrome b5 Inhibits Electron Transfer from NADPH-Cytochrome P450 Reductase to Ferric Cytochrome P450 2B4. J. Biol. Chem. 2008, 283, 5217–5225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NOBLE, M.A.; MILES, C.S.; CHAPMAN, S.K.; LYSEK, D.A.; MACKAY, A.C.; REID, G.A.; HANZLIK, R.P.; MUNRO, A.W. Roles of key active-site residues in flavocytochrome P450 BM3. Biochem. J. 1999, 339, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Li, H.; Poulos, T.L. Structural basis for effector control and redox partner recognition in cytochrome P450. Science 2013, 340, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Pearl, N.M.; Wilcoxen, J.; Im, S.; Kunz, R.; Darty, J.; Britt, R.D.; Ragsdale, S.W.; Waskell, L. Protonation of the Hydroperoxo Intermediate of Cytochrome P450 2B4 Is Slower in the Presence of Cytochrome P450 Reductase Than in the Presence of Cytochrome b5. Biochemistry 2016, 55, 6558–6567. [Google Scholar] [CrossRef] [PubMed]
- Hamdane, D.; Xia, C.; Im, S.-C.; Zhang, H.; Kim, J.-J.P.; Waskell, L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009, 284, 11374–11384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridges, A.; Gruenke, L.; Chang, Y.T.; Vakser, I.A.; Loew, G.; Waskell, L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 1998, 273, 17036–17049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nash, T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem. J. 1953, 55, 416–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saribas, A.S.; Gruenke, L.; Waskell, L. Overexpression and Purification of the Membrane-Bound Cytochrome P450 2B4. Protein Expr. Purif. 2001, 21, 303–309. [Google Scholar] [CrossRef] [PubMed]
| λ= 450 nm | ||||
|---|---|---|---|---|
| Enzyme | k1obs (s−1) (amplitude%) | k2obs (s−1) (amplitude%) | k3obs (s−1) (amplitude%) | R2 |
| WT CPR | 9.93 ± 0.17 (82.8 ± 0.2) | 0.54 ± 0.014 (17.2 ± 0.2) | 0.9951 | |
| ΔG141 | 9.12 ± 0.10 (7.2 ± 0.3) | 0.03 ± 0.001 (89.2 ± 0.6) | 0.01 ± 0.001 (3.6 ± 0.2) | 0.9994 |
| ΔG141/E142N | 16.29 ± 0.31 (60.1 ± 0.8) | 0.03 ± 0.001 (39.9 ± 0.8) | 0.9938 | |
| ΔG143 | 3.34 ± 0.10 (81.8 ± 0.1) | 0.11 ± 0.01 (18.2 ± 0.1) | 0.9943 | |
| Full-Length CYPOR Reductase Mutant/Wild-Type | Molar Ratio P450:CYPOR:Cyt b5 | Benzphetamine Nmol of CH2O Produced/s/nmol CYPOR |
|---|---|---|
| WT | 1:1:0 | 0.68 ± 0.01 |
| 1:1:0.5 | 0.82 ± 0.04 | |
| 1:1:1 | 0.73 ± 0.04 | |
| 1:1:1.5 | 0.63 ± 0.01 | |
| 1:1:2 | 0.52 ± 0.01 | |
| 1:1:5 | 0.26 ± 0.02 | |
| ΔG141 | 1:1:0 | 0.004 ± 0.001 |
| 1:1:0.5 | 0.01 ± 0.003 | |
| 1:1:1 | No activity | |
| 1:1:1.5 | No activity | |
| 1:1:2 | No activity | |
| ΔG141/E142N | 1:1:0 | 0.001 ± 0.0006 |
| 1:1:0.5 | 0.01 ± 0.003 | |
| 1:1:1 | No activity | |
| 1:1:1.5 | No activity | |
| 1:1:2 | No activity | |
| ΔG143 | 1:1:0 | 0.08 ± 0.005 |
| 1:1:0.5 | 0.11 ± 0.003 | |
| 1:1:1 | 0.07 ± 0.011 | |
| 1:1:1.5 | 0.05 ± 0.002 | |
| 1:1:2 | 0.03 ± 0.001 |
| Mutant | Sequence |
|---|---|
| ΔG141 (Forward) | 5′-TGC ATG GCC ACA TAC --- GAG GGC GAC C-3′ |
| ΔG141/E142N (Forward) | 5′-TGC ATG GCC ACA TAC --- AAC GGC GAC C-3′ |
| ΔG143 (Forward) | 5′-ATG GCC ACA TAC GGA GAG --- GAC CCC A-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rwere, F.; Im, S.; Waskell, L. The FMN “140s Loop” of Cytochrome P450 Reductase Controls Electron Transfer to Cytochrome P450. Int. J. Mol. Sci. 2021, 22, 10625. https://doi.org/10.3390/ijms221910625
Rwere F, Im S, Waskell L. The FMN “140s Loop” of Cytochrome P450 Reductase Controls Electron Transfer to Cytochrome P450. International Journal of Molecular Sciences. 2021; 22(19):10625. https://doi.org/10.3390/ijms221910625
Chicago/Turabian StyleRwere, Freeborn, Sangchoul Im, and Lucy Waskell. 2021. "The FMN “140s Loop” of Cytochrome P450 Reductase Controls Electron Transfer to Cytochrome P450" International Journal of Molecular Sciences 22, no. 19: 10625. https://doi.org/10.3390/ijms221910625
APA StyleRwere, F., Im, S., & Waskell, L. (2021). The FMN “140s Loop” of Cytochrome P450 Reductase Controls Electron Transfer to Cytochrome P450. International Journal of Molecular Sciences, 22(19), 10625. https://doi.org/10.3390/ijms221910625







