Trop2 Forms a Stable Dimer with Significant Structural Differences within the Membrane-Distal Region as Compared to EpCAM
Abstract
:1. Introduction
2. Results and Discussion
2.1. Protein Sample Preparation for Crystallization
2.2. Trop2 Ectodomain Structure Overview and Molecular Assemblies
2.2.1. Structure Determination
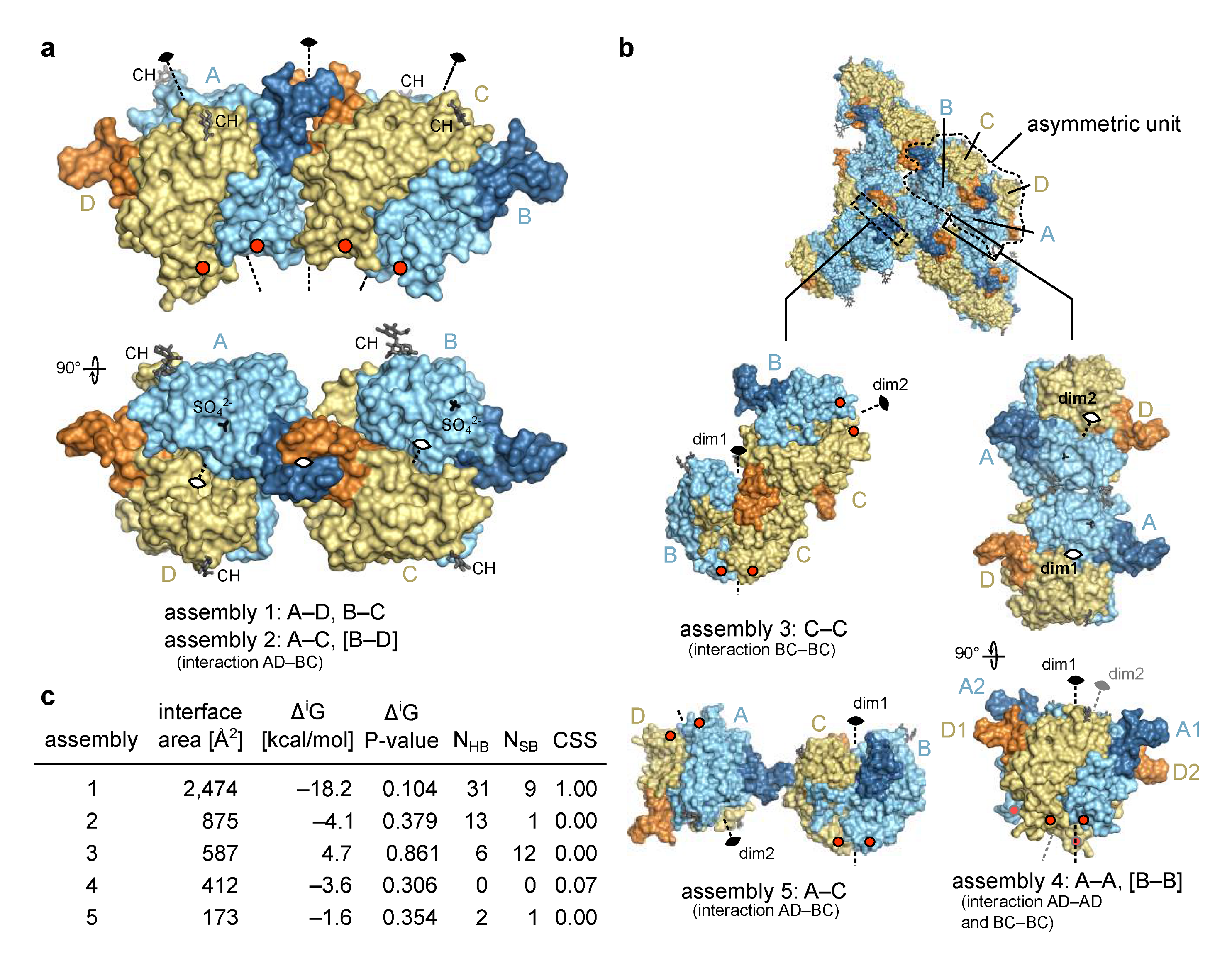
2.2.2. Molecular Assemblies
2.2.3. General Structural Features of the Trop2 Ectodomain
2.3. Trop2 Ectodomain Dimer and It’s Implications in Trop2 Proteolytic Cleavage
2.4. Trop2 and EpCAM Significantly Differ in Their Membrane-Distal Regions
3. Materials and Methods
3.1. Expression and Purification of Trop2 Ectodomain for Crystallization
3.2. N-Terminal Sequencing and Sequence Analysis
3.3. Crystallization
3.4. Data Collection and Structure Solving
3.5. Structure Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stepan, L.; Trueblood, E.S.; Hale, K.; Babcook, J.; Borges, L.; Sutherland, C.L. Expression of trop2 cell surface glycoprotein in normal and tumor tissues. J. Histochem. Cytochem. 2011, 59, 701–710. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.; Lawson, D.A.; Cheng, D.; Sun, W.; Garraway, I.P.; Witte, O.N. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl. Acad. Sci. USA 2008, 105, 20882–20887. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhu, Z.; Wang, H.; Li, F.; Du, X.; Ma, R.Z. Trop2 regulates the proliferation and differentiation of murine compact-bone derived MSCs. Int. J. Oncol. 2013, 43, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Lenárt, S.; Lenárt, P.; Šmarda, J.; Remšík, J.; Souček, K.; Beneš, P. Trop2: Jack of all trades, master of none. Cancers 2020, 12, 3328. [Google Scholar] [CrossRef]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavšič, M.; Gunčar, G.; Djinovic-Carugo, K.; Lenarčič, B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat. Commun. 2014, 5, 4764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casaletto, J.B.; Geddie, M.L.; Abu-Yousif, A.O.; Masson, K.; Fulgham, A.; Boudot, A.; Maiwald, T.; Kearns, J.D.; Kohli, N.; Su, S.; et al. MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM. Proc. Natl. Acad. Sci. USA 2019, 116, 7533–7542. [Google Scholar] [CrossRef] [Green Version]
- Gaber, A.; Lenarčič, B.; Pavšič, M. Current view on EpCAM structural biology. Cells 2020, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Linnenbach, A.J.; Seng, B.A.; Wu, S.; Robbins, S.; Scollon, M.; Pyrc, J.J.; Druck, T.; Huebner, K. Retroposition in a family of carcinoma-associated antigen genes. Mol. Cell. Biol. 1993, 13, 1507–1515. [Google Scholar] [CrossRef] [Green Version]
- Gaber, A.; Kim, S.J.; Kaake, R.M.; Benčina, M.; Krogan, N.; Šali, A.; Pavšič, M.; Lenarčič, B. EpCAM homo-oligomerization is not the basis for its role in cell-cell adhesion. Sci. Rep. 2018, 8, 13269. [Google Scholar] [CrossRef]
- Vidmar, T.; Pavšič, M.; Lenarčič, B. Biochemical and preliminary X-ray characterization of the tumor-associated calcium signal transducer 2 (Trop2) ectodomain. Protein Expr. Purif. 2013, 91, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Hachmeister, M.; Bobowski, K.D.; Hogl, S.; Dislich, B.; Fukumori, A.; Eggert, C.; Mack, B.; Kremling, H.; Sarrach, S.; Coscia, F.; et al. Regulated intramembrane proteolysis and degradation of murine epithelial cell adhesion molecule mEpCAM. PLoS ONE 2013, 8, e71836. [Google Scholar] [CrossRef] [PubMed]
- Tsaktanis, T.; Kremling, H.; Pavšič, M.; von Stackelberg, R.; Mack, B.; Fukumori, A.; Steiner, H.; Vielmuth, F.; Spindler, V.; Huang, Z.; et al. Cleavage and cell adhesion properties of human epithelial cell adhesion molecule (HEPCAM). J. Biol. Chem. 2015, 290, 24574–24591. [Google Scholar] [CrossRef] [Green Version]
- Žagar, T.; Pavšič, M.; Gaber, A. Destabilization of EpCAM dimer is associated with increased susceptibility towards cleavage by TACE. PeerJ 2021, 9, e11484. [Google Scholar] [CrossRef] [PubMed]
- Stoyanova, T.; Goldstein, A.; Cai, H.; Drake, J.; Huang, J.; Witte, O.N. Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via -catenin signaling. Genes Dev. 2012, 26, 2271–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maetzel, D.; Denzel, S.; Mack, B.; Canis, M.; Went, P.T.; Benk, M.; Kieu, C.; Papior, P.; Baeuerle, P.A.; Munz, M.; et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009, 11, 162–171. [Google Scholar] [CrossRef]
- Zhao, W.; Jia, L.; Kuai, X.; Tang, Q.; Huang, X.; Yang, T.; Qiu, Z.; Zhu, J.; Huang, J.; Huang, W.; et al. The role and molecular mechanism of Trop2 induced epithelial-mesenchymal transition through mediated β-catenin in gastric cancer. Cancer Med. 2019, 8, 1135–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, A.; Goldenberg, D.M.; Stein, R. The epithelial/carcinoma antigen EGP-1, recognized by monoclonal antibody RS7–3G11, is phosphorylated on serine 303. Int. J. Cancer 1995, 62, 472–479. [Google Scholar] [CrossRef]
- Pavšič, M.; Ilc, G.; Vidmar, T.; Plavec, J.; Lenarčič, B. The cytosolic tail of the tumor marker protein Trop2—a structural switch triggered by phosphorylation. Sci. Rep. 2015, 5, 10324. [Google Scholar] [CrossRef]
- Wu, C.-J.; Lu, M.; Feng, X.; Nakato, G.; Udey, M.C. Matriptase cleaves EpCAM and trop2 in keratinocytes, destabilizing both proteins and associated claudins. Cells 2020, 9, 1027. [Google Scholar] [CrossRef]
- Mueller, J.L.; McGeough, M.D.; Peña, C.A.; Sivagnanam, M. Functional consequences of EpCam mutation in mice and men. Am. J. Physiol. Liver Physiol. 2014, 306, G278–G288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivagnanam, M.; Mueller, J.L.; Lee, H.; Chen, Z.; Nelson, S.F.; Turner, D.; Zlotkin, S.H.; Pencharz, P.B.; Ngan, B.; Libiger, O.; et al. Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology 2008, 135, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Nakato, G.; Morimura, S.; Lu, M.; Feng, X.; Wu, C.; Udey, M.C. Amelioration of congenital tufting enteropathy in EpCAM (TROP1)-deficient mice via. heterotopic expression of trop2 in intestinal epithelial cells. Cells 2020, 9, 1847. [Google Scholar] [CrossRef]
- Trerotola, M.; Guerra, E.; Ali, Z.; Aloisi, A.L.; Ceci, M.; Simeone, P.; Acciarito, A.; Zanna, P.; Vacca, G.; D’Amore, A.; et al. Trop-2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis. Neoplasia 2021, 23, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.; Schinke, H.; Luxenburger, E.; Kranz, G.; Shakhtour, J.; Libl, D.; Huang, Y.; Gaber, A.; Pavšič, M.; Lenarčič, B.; et al. EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial-mesenchymal transition through modulation of phospho-ERK1/2 in head and neck cancers. PLoS Biol. 2018, 16, e2006624. [Google Scholar] [CrossRef]
- Lin, J.; Wu, Y.; Wu, J.; Lin, T.; Wu, C.-T.; Chang, Y.-L.; Jou, Y.; Hong, T.; Yang, P. TROP2 is epigenetically inactivated and modulates IGF-1R signalling in lung adenocarcinoma. EMBO Mol. Med. 2012, 4, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Sin, S.T.; Li, Y.; Liu, M.; Ma, S.; Guan, X.-Y. TROP-2 exhibits tumor suppressive functions in cervical cancer by dual inhibition of IGF-1R and ALK signaling. Gynecol. Oncol. 2019, 152, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Jones, L.; Lim, S.; Maher, C.A.; Adkins, D.; Lewis, J.; Kimple, R.J.; Fertig, E.; Chung, C.H.; Herrlich, A.; et al. Loss of Trop2 causes ErbB3 activation through a neuregulin-1-dependent mechanism in the mesenchymal subtype of HNSCC. Oncotarget 2014, 5, 9281–9294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trerotola, M.; Jernigan, D.L.; Liu, Q.; Siddiqui, J.; Fatatis, A.; Languino, L.R. Trop-2 promotes prostate cancer metastasis by modulating β1 integrin functions. Cancer Res. 2013, 73, 3155–3167. [Google Scholar] [CrossRef] [Green Version]
- Trerotola, M.; Ganguly, K.K.; Fazli, L.; Fedele, C.; Lu, H.; Dutta, A.; Liu, Q.; De Angelis, T.; Riddell, L.W.; Riobo, N.A.; et al. Trop-2 is up-regulated in invasive prostate cancer and displaces FAK from focal contacts. Oncotarget 2015, 6, 14318–14328. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.M.; Speicher, D.W. Determination of disulfide bond assignments and N-glycosylation sites of the human gastrointestinal carcinoma antigen GA733-2 (CO17-1A, EGP, KS1-4, KSA, and Ep-CAM). J. Biol. Chem. 2001, 276, 5804–5813. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2017, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010, 31, 133–143. [Google Scholar] [CrossRef]
- Capitani, G.; Duarte, J.M.; Baskaran, K.; Bliven, S.; Somody, J.C. Understanding the fabric of protein crystals: Computational classification of biological interfaces and crystal contacts. Bioinformatics 2016, 32, 481–489. [Google Scholar] [CrossRef]
- Elez, K.; Bonvin, A.M.J.J.; Vangone, A. Biological vs. crystallographic protein interfaces: An overview of computational approaches for their classification. Crystals 2020, 10, 114. [Google Scholar] [CrossRef] [Green Version]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Hua, P.; Lou, Y.; Li, Z.; Jia, M.; Jing, Y.; Cai, M.; Wang, H.; Tong, T.; Gao, J. Mechanistic insights into trop2 clustering on lung cancer cell membranes revealed by super-resolution imaging. ACS Omega 2020, 5, 32456–32465. [Google Scholar] [CrossRef]
- Herget, S.; Ranzinger, R.; Maass, K.; Lieth, C.-W. GlycoCT—A unifying sequence format for carbohydrates. Carbohydr. Res. 2008, 343, 2162–2171. [Google Scholar] [CrossRef]
- Shi, X.; Jarvis, D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 2007, 8, 1116–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, M.; Park, D.-Y.; Kim, Y.; Lee, K.-J.; Lu, Z.; Ko, K.; Choo, Y.K.; Han, Y.S.; Ahn, M.-H.; Oh, D.-B.; et al. Characterization of N-glycan structures and biofunction of anti-colorectal cancer monoclonal antibody CO17-1A produced in baculovirus-insect cell expression system. J. Biosci. Bioeng. 2010, 110, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Munz, M.; Fellinger, K.; Hofmann, T.; Schmitt, B.; Gires, O. Glycosylation is crucial for stability of tumour and cancer stem cell antigen EpCAM. Front. Biosci. 2008, 13, 5195–5201. [Google Scholar] [CrossRef] [Green Version]
- Kamble, P.R.; Rane, S.; Breed, A.A.; Joseph, S.; Mahale, S.D.; Pathak, B.R. Proteolytic cleavage of Trop2 at Arg87 is mediated by matriptase and regulated by Val194. FEBS Lett. 2020, 594, 3156–3169. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Feng, X.; Lu, M.; Morimura, S.; Udey, M.C. Matriptase-mediated cleavage of EpCAM destabilizes claudins and dysregulates intestinal epithelial homeostasis. J. Clin. Investig. 2017, 127, 623–634. [Google Scholar] [CrossRef]
- Horton, R.M.; Hunt, H.D.; Ho, S.N.; Pullen, J.K.; Pease, L.R. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 1989, 77, 61–68. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Kabsch, W. Research papers XDS research papers. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.; Winn, M.D.; Storoni, L.C.; Read, R. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Matthews, B. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Kantardjieff, K.A.; Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003, 12, 1865–1871. [Google Scholar] [CrossRef]
- Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 1002–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowtan, K.D. Completion of autobuilt protein models using a database of protein fragments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 328–335. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.; Cowtan, K.D. Features and development of coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- The PyMOL Molecular Graphics System, version 2.4.0; Schrödinger, LLC: New York, NY, USA, 2021.
- Shindyalov, I.N.; Bourne, P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. Des. Sel. 1998, 11, 739–747. [Google Scholar] [CrossRef]




| PDB ID | 7PEE |
|---|---|
| Data collection | |
| X-ray source and beamline | Elettra Synchrotron, XRD2 (11.2C) |
| Wavelength (Å) | 0.9789 |
| Space group | P4322 |
| Cell dimensions | |
| a, b, c (Å) | 145.08, 145.08, 217.77 |
| α, β, γ (°) | 90, 90, 90 |
| Data statistics | |
| Resolution range (Å) a | 48.36–2.81 (2.91–2.81) |
| Total no. of reflections a | 376,492 (36,911) |
| No. of unique reflections a | 57,144 (5604) |
| Mean I/σ(I) a | 15.27 (1.73) |
| Rmerge (%) a,b | 11.88 (127.2) |
| CC1/2 a,b | 0.997 (0.692) |
| Completeness (%) a | 99.59 (99.48) |
| Redundancy a | 6.6 (6.6) |
| Number of atoms | |
| Total | 7620 |
| Protein/Water/Ligands | 7476/30/114 |
| Refinement statistics | |
| Rwork/Rfree (%) c | 23.94 (26.30) |
| Root-mean-square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.07 |
| Ramachandran plot | |
| Favored/Allowed/Outliers (%) d | 95.75/4.25/0.00 |
| Rotamer outliers (%) d | 1.71 |
| B-factor | |
| Average | 75.80 |
| Protein/Ligands/Solvent | 75.12/125.23/57.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavšič, M. Trop2 Forms a Stable Dimer with Significant Structural Differences within the Membrane-Distal Region as Compared to EpCAM. Int. J. Mol. Sci. 2021, 22, 10640. https://doi.org/10.3390/ijms221910640
Pavšič M. Trop2 Forms a Stable Dimer with Significant Structural Differences within the Membrane-Distal Region as Compared to EpCAM. International Journal of Molecular Sciences. 2021; 22(19):10640. https://doi.org/10.3390/ijms221910640
Chicago/Turabian StylePavšič, Miha. 2021. "Trop2 Forms a Stable Dimer with Significant Structural Differences within the Membrane-Distal Region as Compared to EpCAM" International Journal of Molecular Sciences 22, no. 19: 10640. https://doi.org/10.3390/ijms221910640
APA StylePavšič, M. (2021). Trop2 Forms a Stable Dimer with Significant Structural Differences within the Membrane-Distal Region as Compared to EpCAM. International Journal of Molecular Sciences, 22(19), 10640. https://doi.org/10.3390/ijms221910640






