Genome-Wide Identification and Characterization of Polygalacturonase Gene Family in Maize (Zea mays L.)
Abstract
:1. Introduction
2. Results
2.1. Identification of PG Genes from Maize Genome
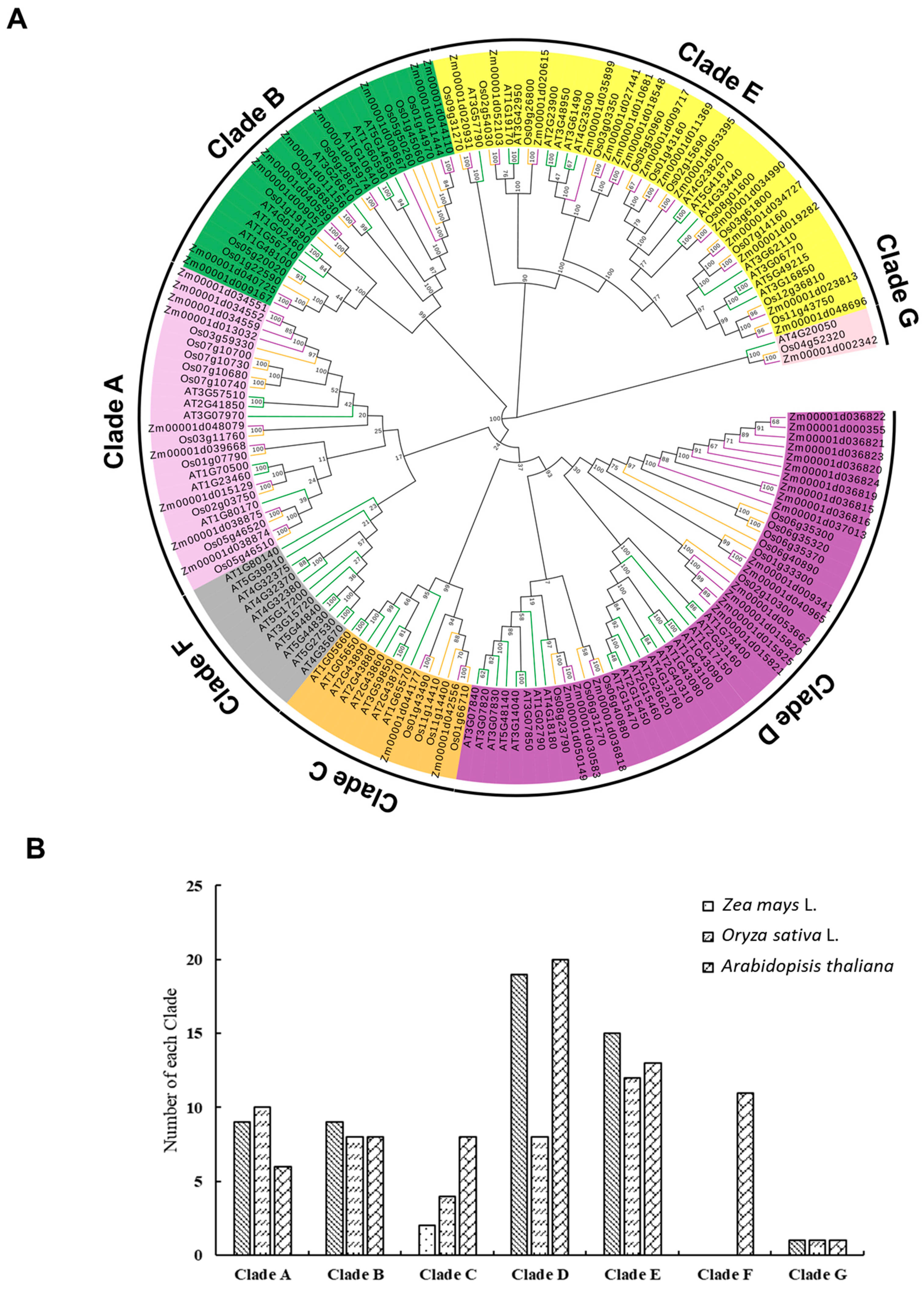
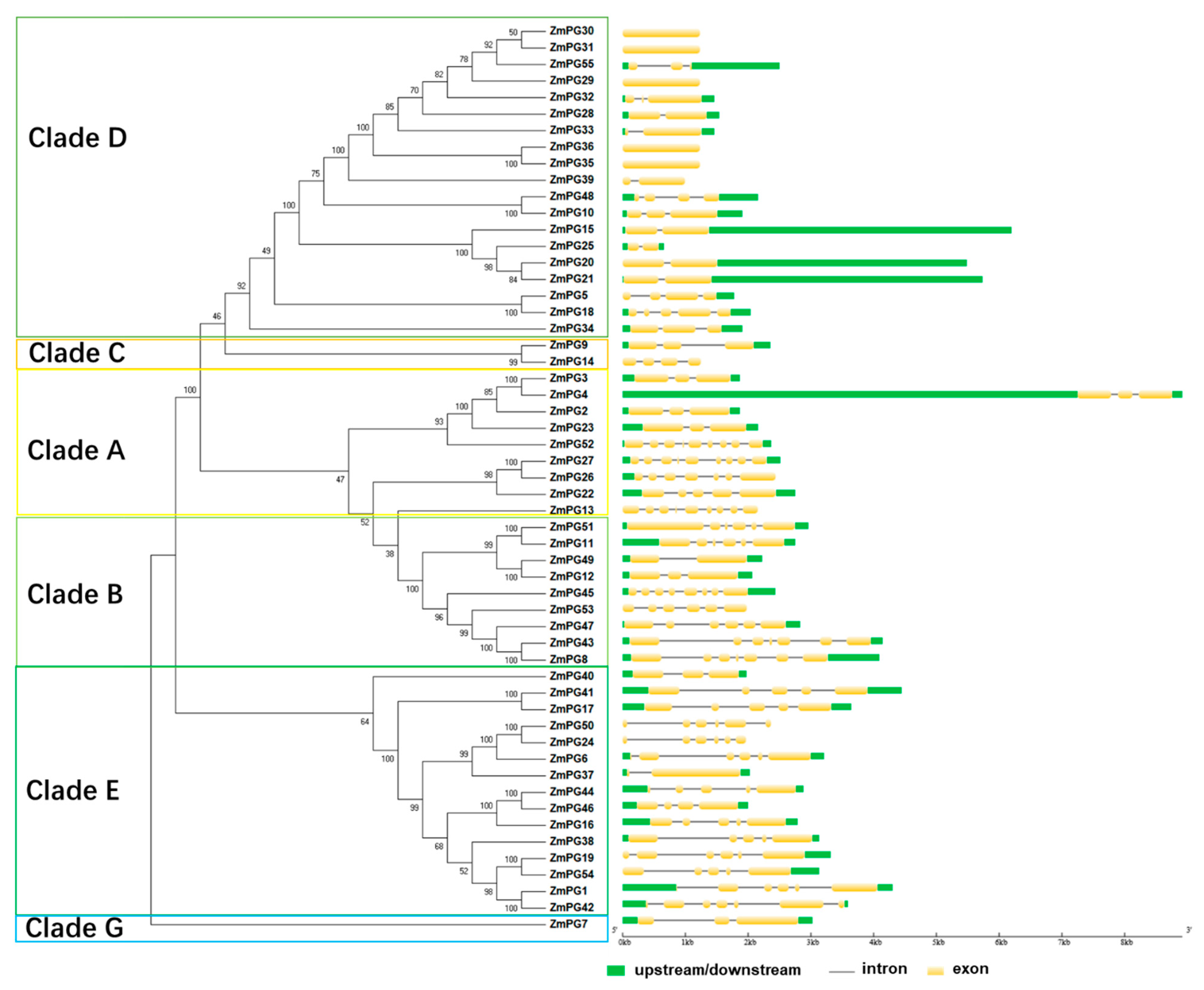
2.2. Phylogenetic Analysis of ZmPGs
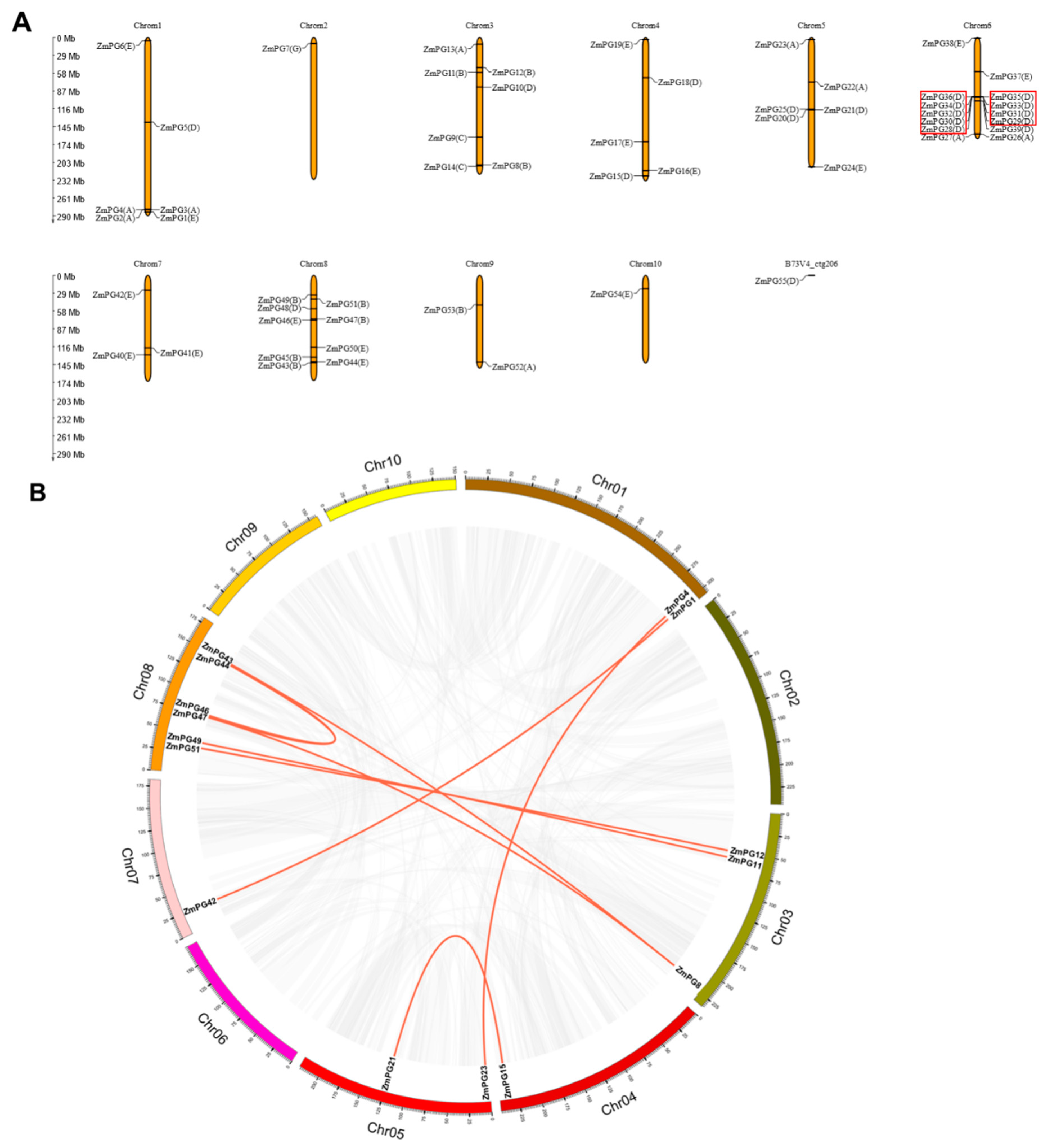
2.3. Collinearity and Amino Acid Substitution Selection Pressure Analyses
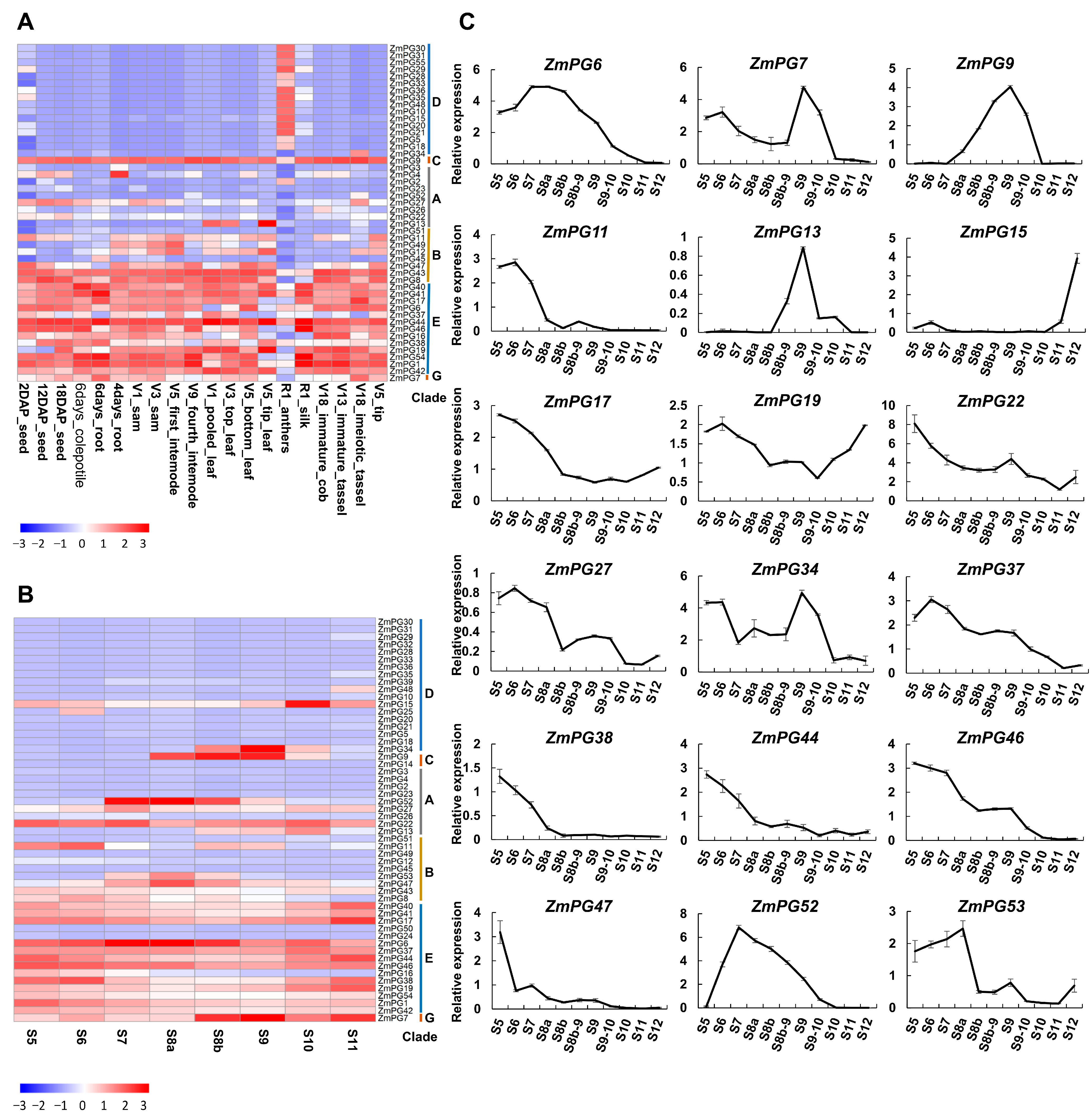
2.4. Expression Analysis of Maize PG Genes in Different Tissues and Developmental Anthers
2.5. Validation of the PG Gene Expression in Developmental Anthers via RT-qPCR
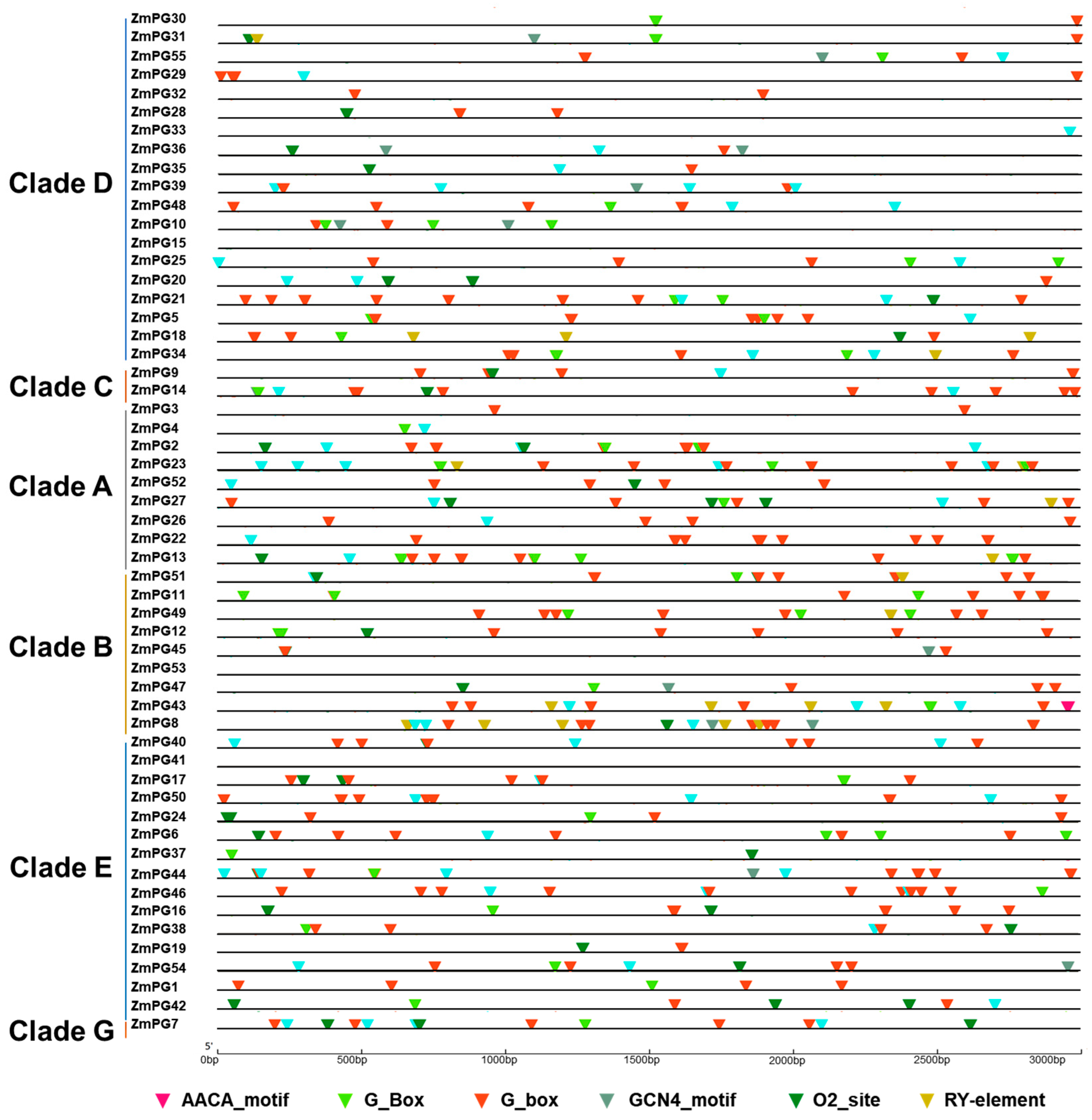
2.6. Cis-Regulatory Motif Analysis of ZmPG Genes
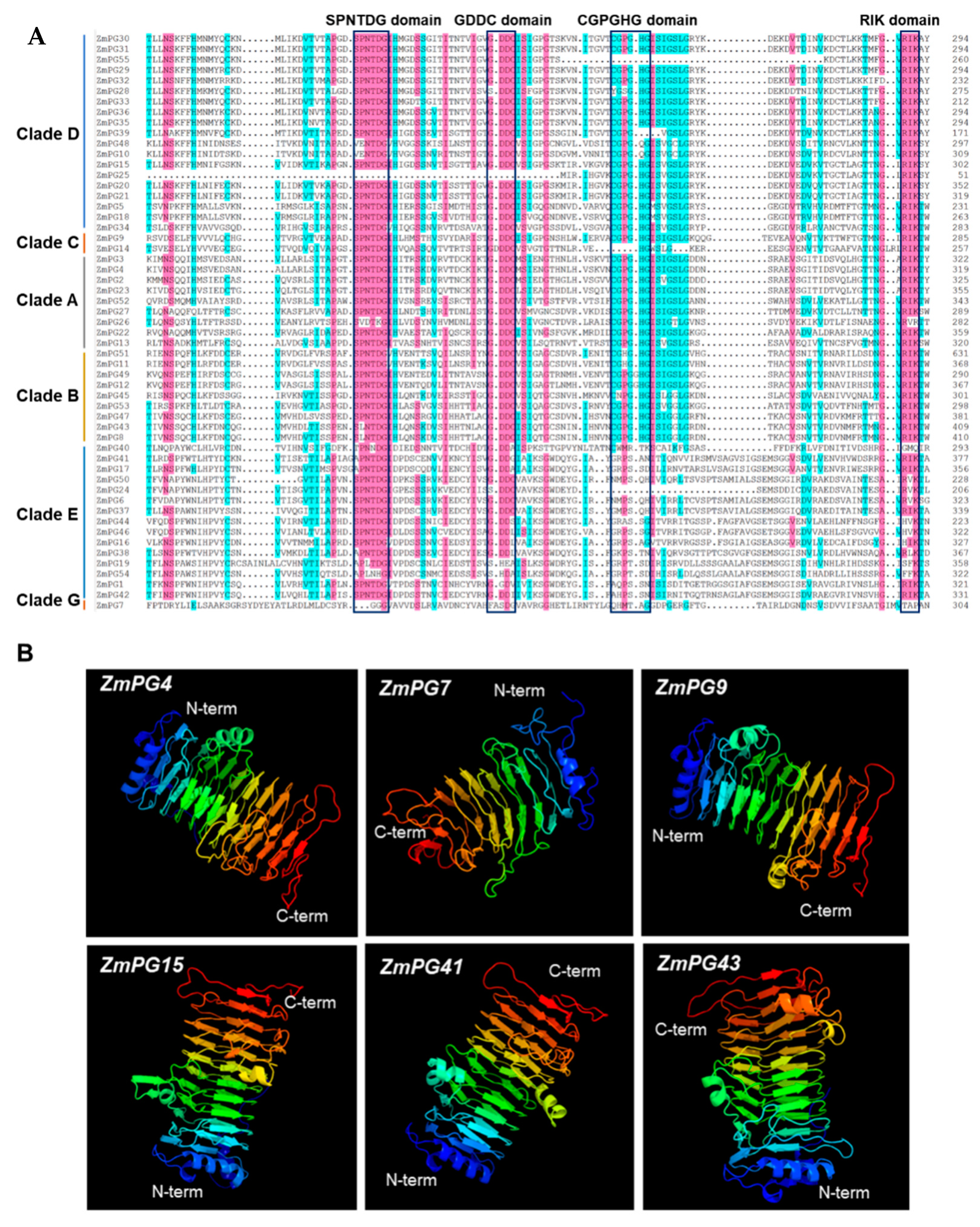
2.7. Conserved Motif and Structure Prediction of the Maize PG Proteins
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of PG Genes in Maize
4.2. Phylogenetic Analysis and Chromosomal Location of Maize PG Genes
4.3. Gene Structure and Conserved Motif Analysis
4.4. Collinearity Analysis and Selective Pressure for Duplicated Genes
4.5. Cis-Elements Analysis of Maize PG Gene Promoters
4.6. Gene Model Analysis and the Functional Connection Network Analysis
4.7. Expression Analysis of Maize PG Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, J. The pectin lyases in Arabidopsis thaliana: Evolution, selection and expression profiles. PLoS ONE 2012, 7, e46944. [Google Scholar] [CrossRef]
- Lang, C.; Dornenburg, H. Perspectives in the biological function and the technological application of polygalacturonases. Appl. Microbiol. Biotechnol. 2000, 53, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Kwon, S.J.; Kim, N.S. Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genom. 2010, 32, 570–577. [Google Scholar] [CrossRef]
- Rcxovabenkova, L. Evidence for the role of carboxyl groups in activity of endopolygalacturonase of aspelus-niger chemical modification by carbodiimide reagent. Collect. Czech. Chem. Commun. 1990, 55, 1389–1395. [Google Scholar] [CrossRef]
- Rao, M.N.; Kembhavi, A.A.; Pant, A. Implication of tryptophan and histidine in the active site of endo-polygalacturonase from Aspergillus ustus: Elucidation of the reaction mechanism. BBA Protein Struct. Mol. Enzymol. 1996, 1296, 167–173. [Google Scholar] [CrossRef]
- Park, K.C.; Kwon, S.J.; Kim, P.H.; Bureau, T.; Kim, N.S. Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 2008, 51, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, Y.; Liang, Y.; Anderson, C.T.; Cao, J. A Profusion of molecular scissors for pectins: Classification, expression, and functions of plant polygalacturonases. Front. Plant Sci. 2018, 9, 1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirsch, R.; Gramzow, L.; Theiben, G.; Siegfried, B.D.; Ffrench-Constant, R.H.; Heckel, D.G.; Pauchet, Y. Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: Key events in the evolution of herbivory in beetles. Insect Biochem. Mol. Biol. 2014, 52, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.; Pickersgill, R. The architecture of parallel β-helices and related folds. Prog. Biophys. Mol. Biol. 2001, 77, 111–175. [Google Scholar] [CrossRef]
- Chothia, C.; Hubbard, T.; Brenner, S.; Barns, H.; Murzin, A. Protein folds in the all-β and all-α classes. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 597–627. [Google Scholar] [CrossRef]
- Abbott, D.W.; Boraston, A.B. The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 2007, 368, 1215–1222. [Google Scholar] [CrossRef]
- Choi, J.K.; Lee, B.H.; Chae, C.H.; Shin, W. Computer modeling of the rhamnogalacturonase-“hairy” pectin complex. Proteins 2004, 55, 22–33. [Google Scholar] [CrossRef]
- Brummell, D.A.; Harpster, M.H. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 2001, 47, 311–340. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.H.; Thoma, S.; Li, W.H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, C.; Somerville, C.; Anderson, C.T. Polygalacturonase involved in expression functions in cell elongation and flower development in Arabidopsis. Plant Cell 2014, 26, 1018–1035. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.Y.; Somerville, C.R. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 1998, 15, 79–88. [Google Scholar] [CrossRef]
- Zhang, A.; Chen, Q.; Huang, L.; Qiu, L.; Cao, J. Cloning and expression of an anther-abundant polygalacturonase gene BcMF17 from Brassica campestris ssp. chinensis. Plant Mol. Biol. Rep. 2011, 29, 943–951. [Google Scholar] [CrossRef]
- Zhang, A.; Chen, Q.; Huang, L.; Qiu, L.; Cao, J. Isolation and characterization of an anther-specific polygalacturonase gene, BcMF16, in Brassica campestris ssp. chinensis. Plant Mol. Biol. Rep. 2012, 30, 330–338. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, L.; Liu, T.; Yu, X.; Cao, J. Functional analysis of a pollen-expressed polygalacturonase gene BcMF6 in Chinese cabbage (Brassica campestris L. ssp. chinensis Makino). Plant Cell Rep. 2008, 27, 1207–1215. [Google Scholar] [CrossRef]
- Gorguet, B.; Schipper, D.; van Lammeren, A.; Visser, R.F.; van Heusden, A.W. Ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor. Appl. Genet. 2009, 118, 1199–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quesada, M.A.; Blanco-Portales, R.; Pose, S.; Garcia-Gago, J.A.; Jimenez-Bermudez, S.; Munoz-Serrano, A.; Caballero, J.L.; Pliego-Alfaro, F.; Mercado, J.A.; Munoz-Blanco, J. Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in Strawberry Fruit Softening. Plant Physiol. 2009, 150, 1022–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Wang, L.; Wang, W.; Zhou, H.; Ma, B.Q.; Zheng, H.Y.; Fang, T.; Ogutu, C.; Vimolmangkang, S.; Han, Y.P. Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 2016, 67, 1993–2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dautt-Castro, M.; Lopez-Virgen, A.G.; Ochoa-Leyva, A.; Contreras-Vergara, C.A.; Sortillon-Sortillon, A.P.; Martinez-Tellez, M.A.; Gonzalez-Aguilar, G.A.; Casas-Flores, J.S.; Sanudo-Barajas, A.; Kuhn, D.N.; et al. Genome-wide identification of mango (Mangifera indica L.) polygalacturonases: Expression analysis of family members and total enzyme activity during fruit ripening. Front. Plant Sci. 2019, 10, 969. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.; Zheng, T.; Fang, J. Genome-wide identification and expression profiling of the polygalacturonase (PG) and pectin methylesterase (PME) genes in Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.L.; Yang, X.T.; Yang, Z.Q.; Niu, F.Q.; Chen, Y.R.; Zhang, L.L.; Song, X.Y. Comprehensive analysis of polygalacturonase gene family highlights candidate genes related to pollen development and male fertility in wheat (Triticum aestivum L.). Planta 2020, 252, 31. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Cui, J.; Lyu, M.; Xu, L.; Cao, J. A comparative analysis of the evolution, expression, and cis-regulatory element of polygalacturonase genes in grasses and dicots. Funct. Integr. Genomics. 2016, 16, 641–656. [Google Scholar] [CrossRef]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197, 1353–1365. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Shen, X.; Dong, H.; Lyu, M.; Xu, L.; Ma, Z.; Liu, T.; Cao, J. Dissecting the complex molecular evolution and expression of polygalacturonase gene family in Brassica rapa ssp. chinensis. Plant Mol. Biol. 2015, 89, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.F.; Sun, X.; Shi, X.Y.; Zhai, H.; Tian, C.G.; Kong, F.J.; Liu, B.H.; Yuan, X.H. A global analysis of the polygalacturonase gene family in soybean (Glycine max). PLoS ONE 2016, 11, e0163012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y.; Cao, J.; Zhu, Z.; Yu, Y. Genome-wide identification and analysis of polygalacturonase genes in solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.J.; Chen, M.Y.; Zhao, T.T.; Han, F.; Zhang, Q.; Liu, X.L.; Jiang, C.Y.; Zhong, C.H. Genome-wide identification and expression analysis of polygalacturonase gene family in Kiwifruit (Actinidia chinensis) during fruit softening. Plants 2020, 9, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, T.; Huang, X.G.; Pan, X.T.; Zhang, J.; Xie, R.J. Genome-wide identification and expression analysis of citrus fruitlet abscission-related polygalacturonase genes. 3 Biotech. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Shao, H.X.; Fan, S.; Ma, J.J.; Zhang, D.; Han, M.Y. Identification and phylogenetic analysis of the polygalacturonase gene family in Apple. Hortic. Plant J. 2016, 2, 241–252. [Google Scholar] [CrossRef]
- He, Y.; Song, S.; Li, M.Y.; Bo, W.H.; Li, Y.Y.; Cao, M.; Wang, A.; Li, G.Q.; Liu, X.Y.; Pang, X.M. Identification and expression analysis of the polygalacturonase Gene Family in Chinese Jujube (Ziziphus jujuba Mill.). Mol. Plant Breed. 2021, 19, 1442–1450. [Google Scholar] [CrossRef]
- Qian, M.; Zhang, Y.; Yan, X.Y.; Han, M.Y.; Li, J.J.; Li, F.; Li, F.R.; Zhang, D.; Zhao, C.P. Identification and expression analysis of polygalacturonase family members during peach fruit softening. Int. J. Mol. Sci. 2016, 17, 1933. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.J.; Liang, Y.; Lv, M.M.; Wu, J.; Lu, G.; Cao, J.S. Genome-wide identification and characterization of polygalacturonase genes in cucumis sativus and citrullus lanatus. Plant Physiol. Bioch. 2014, 74, 263–275. [Google Scholar] [CrossRef]
- Hadfield, K.A.; Rose, J.K.; Yaver, D.S.; Berka, R.M.; Bennett, A.B. Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening-associated pectin disassembly. Plant Physiol. 1998, 117, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Wang, X.; Wang, H.; Xin, H.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013, 9, e1003790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.; Lou, Y.; Shen, S.Y.; Wang, S.H.; Zhou, L.; Wang, J.; Liu, X.L.; Xiong, S.X.; Han, Y.; Zhou, H.S.; et al. A cellular mechanism underlying the restoration of thermo/photoperiod-sensitive genic male sterility. Mol. Plant 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Hase, T.; Toyoda, T.; Shinozaki, K.; Okamoto, M. Origin and evolution of genes related to ABA metabolism and its signaling pathways. J. Plant Res. 2011, 124, 455–465. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Wang, J.; Lin, W.; Li, S.G.; Li, H.; Zhou, J.; Ni, P.X.; Dong, W.; Hu, S.N.; Zeng, C.Q.; et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, 266–281. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Clarke, J.D.; Zhang, Y.; Dong, X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 2001, 14, 1131–1139. [Google Scholar] [CrossRef] [Green Version]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.F.; Zhang, Y.Y.; Zhang, C.J.; Zhang, T.Y.; Hu, T.X.; Ye, J.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Ye, Z.B. Genome-wide analysis of plant-specific dof transcription factor family in tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef]
- Gaut, B.S.; Morton, B.R.; McCaig, B.C.; Clegg, M.T. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 1996, 93, 10274–10279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Dawson, N.L.; Lewis, T.E.; Das, S.; Lees, J.G.; Lee, D.; Ashford, P.; Orengo, C.A.; Sillitoe, I. CATH: An expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017, 45, D289–D295. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tebbutt, S.J.; Rogers, H.J.; Lonsdale, D.M. Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol. Biol. 1994, 25, 283–297. [Google Scholar] [CrossRef]
- Lewis, T.E.; Sillitoe, I.; Dawson, N.; Lam, S.D.; Clarke, T.; Lee, D.; Orengo, C.; Lees, J. Gene3D: Extensive prediction of globular domains in proteins. Nucleic Acids Res. 2018, 46, D435–D439. [Google Scholar] [CrossRef]
- Pickersgill, R.; Smith, D.; Worboys, K.; Jenkins, J. Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J. Biol. Chem. 1998, 273, 24660–24664. [Google Scholar] [CrossRef] [Green Version]
- Van Santen, Y.; Benen, J.A.; Schröter, K.H.; Kalk, K.H.; Armand, S.; Visser, J.; Dijkstra, B.W. 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 1999, 274, 30474–30480. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, Z. Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a beta-trefoil fold. Biosci. Biotechnol. Biochem. 2013, 77, 1363–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, X.; Wu, S.; Li, Z.; Dong, Z.; An, X.; Ma, B.; Tian, Y.; Li, J. Maize genic male-sterility genes and their applications in hybrid breeding: Progress and perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef] [Green Version]
- Farré, D.; Bellora, N.; Mularoni, L.; Messeguer, X.; Albà, M.M. Housekeeping genes tend to show reduced upstream sequence conservation. Genome Biol. 2007, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Freilich, S.; Massingham, T.; Bhattacharyya, S.; Ponsting, H.; Lyons, P.A.; Freeman, T.C.; Thornton, J.M. Relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biol. 2005, 6, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.J.; Huang, J.C.; Jauh, G.Y. Pollen germination and tube growth. In Advances in Botanical Research; Kader, J.-C., Delseny, M., Eds.; Academic Press: New York, NY, USA, 2010; Volume 54, pp. 1–52. [Google Scholar] [CrossRef]
- Brown, S.M.; Crouch, M.L. Characterization of a gene family abundantly expressed in oenothera organensis pollen that shows sequence similarity to polygalacturonase. Plant Cel.l 1990, 2, 263–274. [Google Scholar] [CrossRef] [Green Version]
- Albani, D.; Altosaar, I.; Arnison, P.G.; Fabijanski, S.F. A gene showing sequence similarity to pectin esterase is specifically expressed in developing pollen of Brassica napus. Sequences in its 5′ flanking region are conserved in other pollen-specific promoters. Plant Mol. Biol. 1991, 16, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Sexton, R.; Del Campillo, E.; Duncan, D.; Lewis, L.N. The purification of an anther cellulase (β(1:4)4-glucan hydrolase) from Lathyrus odoratus L. and its relationship to the similar enzyme found in abscission zones. Plant Sci. 1990, 67, 169–176. [Google Scholar] [CrossRef]
- Allen, R.L.; Lonsdale, D.M. Sequence analysis of three members of the maize polygalacturonase gene family expressed during pollen development. Plant Mol. Biol. 1992, 20, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.S.; Allard, S.; Gerster, J.L.; Cass, L.; Simmonds, J. Isolation and characterization of a polygalacturonase gene highly expressed in Brassica napus pollen. Plant Mol. Biol. 1993, 23, 1273–1278. [Google Scholar] [CrossRef]
- John, M.E.; Petersen, M.W. Cotton (Gossypium hirsutum L.) pollen-specific polygalacturonase mRNA: Tissue and temporal specificity of its promoter in transgenic tobacco. Plant Mol. Biol. 1994, 26, 1989–1993. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Cao, J. Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 2013, 15, 249–263. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.B.; Fabian, A.F.; Martin, G.; Charles, E.C.; Luca, R.; Jingyuan, L.; Wilfred, W.N.; William, S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, L.Q.; Guo, K.; Li, Y.; Tu, Y.Y.; Hu, H.Z.; Wang, B.R.; Cui, X.C.; Peng, L.C. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Coles, N.D.; McMullen, M.D.; Balint-Kurti, P.J.; Pratt, R.C.; Holland, J.B. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 2010, 184, 799–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroll, K.W.; Mokaram, N.E.; Pelletier, A.R.; Frankhouser, D.E.; Westphal, M.S.; Stump, P.A.; Stump, C.L.; Bundschuh, R.; Blachly, J.S.; Yan, P. Quality control for RNA-Seq (QuaCRS): An integrated quality control pipeline. Cancer Inform. 2014, 13, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Chromosome | Length (aa) | MW | pI | Subcellular Localization Prediction | Clade |
|---|---|---|---|---|---|---|---|
| ZmPG1 | Zm00001d034727 | 1 | 477 | 51.69 | 6.08 | extracellular space | E |
| ZmPG2 | Zm00001d034559 | 1 | 475 | 51.21 | 8.98 | extracellular space | A |
| ZmPG3 | Zm00001d034552 | 1 | 441 | 47.30 | 8.46 | extracellular space | A |
| ZmPG4 | Zm00001d034551 | 1 | 436 | 46.90 | 8.83 | extracellular space | A |
| ZmPG5 | Zm00001d030583 | 1 | 344 | 37.24 | 9.02 | extracellular space | D |
| ZmPG6 | Zm00001d027441 | 1 | 463 | 49.74 | 6.16 | extracellular space | E |
| ZmPG7 | Zm00001d002342 | 2 | 496 | 51.30 | 6.09 | extracellular space | G |
| ZmPG8 | Zm00001d044110 | 3 | 542 | 58.27 | 8.47 | extracellular space | B |
| ZmPG9 | Zm00001d042556 | 3 | 401 | 41.93 | 8.99 | extracellular space | C |
| ZmPG10 | Zm00001d040965 | 3 | 428 | 44.61 | 5.78 | extracellular space | D |
| ZmPG11 | Zm00001d040725 | 3 | 502 | 53.47 | 5.17 | organelle membrane | B |
| ZmPG12 | Zm00001d040589 | 3 | 499 | 51.60 | 6.42 | extracellular space | B |
| ZmPG13 | Zm00001d039668 | 3 | 437 | 47.02 | 9.14 | chloroplast | A |
| ZmPG14 | Zm00001d044177 | 3 | 287 | 29.77 | 5.97 | extracellular space | C |
| ZmPG15 | Zm00001d053662 | 4 | 418 | 43.92 | 8.96 | extracellular space | D |
| ZmPG16 | Zm00001d053395 | 4 | 451 | 48.17 | 8.82 | extracellular space | E |
| ZmPG17 | Zm00001d052103 | 4 | 495 | 54.11 | 9.05 | plasma membrane | E |
| ZmPG18 | Zm00001d050149 | 4 | 377 | 40.29 | 9.12 | extracellular space | D |
| ZmPG19 | Zm00001d048696 | 4 | 494 | 52.45 | 5.24 | plasma membrane | E |
| ZmPG20 | Zm00001d015825 | 5 | 468 | 49.33 | 8.74 | plasma membrane | D |
| ZmPG21 | Zm00001d015821 | 5 | 435 | 45.61 | 8.62 | plasma membrane | D |
| ZmPG22 | Zm00001d015129 | 5 | 516 | 54.60 | 5.00 | extracellular space | A |
| ZmPG23 | Zm00001d013032 | 5 | 487 | 51.32 | 6.70 | plasma membrane | A |
| ZmPG24 | Zm00001d018548 | 5 | 232 | 25.61 | 4.93 | nucleus | E |
| ZmPG25 | Zm00001d015820 | 5 | 148 | 15.42 | 9.00 | extracellular space | D |
| ZmPG26 | Zm00001d038875 | 6 | 471 | 49.99 | 5.83 | extracellular space | A |
| ZmPG27 | Zm00001d038874 | 6 | 412 | 43.66 | 8.06 | extracellular space | A |
| ZmPG28 | Zm00001d036824 | 6 | 391 | 41.77 | 6.39 | nucleus | D |
| ZmPG29 | Zm00001d036823 | 6 | 410 | 43.44 | 6.95 | extracellular space | D |
| ZmPG30 | Zm00001d036822 | 6 | 410 | 43.44 | 6.59 | extracellular space | D |
| ZmPG31 | Zm00001d036821 | 6 | 410 | 43.47 | 6.59 | extracellular space | D |
| ZmPG32 | Zm00001d036820 | 6 | 348 | 37.09 | 6.92 | extracellular space | D |
| ZmPG33 | Zm00001d036819 | 6 | 328 | 35.47 | 5.98 | extracellular space | D |
| ZmPG34 | Zm00001d036818 | 6 | 403 | 42.51 | 9.14 | extracellular space | D |
| ZmPG35 | Zm00001d036816 | 6 | 410 | 43.28 | 8.44 | extracellular space | D |
| ZmPG36 | Zm00001d036815 | 6 | 410 | 43.23 | 8.44 | extracellular space | D |
| ZmPG37 | Zm00001d035899 | 6 | 486 | 51.74 | 6.30 | extracellular space | E |
| ZmPG38 | Zm00001d034990 | 6 | 493 | 53.18 | 6.01 | plasma membrane | E |
| ZmPG39 | Zm00001d037013 | 6 | 287 | 30.06 | 7.41 | extracellular space | D |
| ZmPG40 | Zm00001d020931 | 7 | 457 | 50.30 | 8.00 | extracellular space | E |
| ZmPG41 | Zm00001d020615 | 7 | 516 | 56.10 | 9.46 | mitochondrion | E |
| ZmPG42 | Zm00001d019282 | 7 | 508 | 54.55 | 6.17 | extracellular space | E |
| ZmPG43 | Zm00001d011444 | 8 | 541 | 58.09 | 8.03 | extracellular space | B |
| ZmPG44 | Zm00001d011369 | 8 | 348 | 37.36 | 5.49 | extracellular space | E |
| ZmPG45 | Zm00001d011156 | 8 | 436 | 46.16 | 9.21 | extracellular space | B |
| ZmPG46 | Zm00001d009717 | 8 | 446 | 46.99 | 8.12 | extracellular space | E |
| ZmPG47 | Zm00001d009667 | 8 | 516 | 54.74 | 6.84 | plasma membrane | B |
| ZmPG48 | Zm00001d009341 | 8 | 416 | 43.05 | 5.83 | anchored component of plasma membrane | D |
| ZmPG49 | Zm00001d009057 | 8 | 423 | 44.15 | 5.69 | extracellular space | B |
| ZmPG50 | Zm00001d010681 | 8 | 283 | 31.20 | 5.99 | nucleus | E |
| ZmPG51 | Zm00001d009167 | 8 | 766 | 79.73 | 9.22 | nucleus | B |
| ZmPG52 | Zm00001d048079 | 9 | 462 | 49.63 | 6.01 | plasma membrane | A |
| ZmPG53 | Zm00001d045974 | 9 | 419 | 43.59 | 6.24 | plasma membrane | B |
| ZmPG54 | Zm00001d023813 | 10 | 461 | 49.31 | 5.25 | anchored component of plasma membrane | E |
| ZmPG55 | Zm00001d000355 | Contig B73V4_ctg206 | 498 | 53.70 | 8.66 | extracellular space | D |
| No. | Sequence | Duplication Type | Ka | Ks | Ka/Ks | Date (Millions of Years Ago) |
|---|---|---|---|---|---|---|
| 1 | ZmPG20 & ZmPG21 | Tandem | 0.007 | 0.06 | 0.109 | 1.833 |
| 2 | ZmPG30 & ZmPG31 | Tandem | 0.002 | 0.017 | 0.115 | 0.521 |
| 3 | ZmPG30 & ZmPG29 | Tandem | 0.005 | 0.049 | 0.098 | 1.488 |
| 4 | ZmPG31 & ZmPG29 | Tandem | 0.007 | 0.056 | 0.122 | 1.684 |
| 5 | ZmPG36 & ZmPG35 | Tandem | 0.005 | 0.078 | 0.06 | 2.375 |
| 6 | ZmPG32 & ZmPG29 | Tandem | 0.018 | 0.135 | 0.133 | 4.089 |
| 7 | ZmPG30 & ZmPG32 | Tandem | 0.02 | 0.144 | 0.14 | 4.371 |
| 8 | ZmPG31 & ZmPG32 | Tandem | 0.023 | 0.135 | 0.167 | 4.1 |
| 9 | ZmPG3 & ZmPG4 | Tandem | 0.025 | 0.103 | 0.245 | 3.122 |
| 10 | ZmPG28 & ZmPG29 | Tandem | 0.027 | 0.233 | 0.117 | 7.056 |
| 11 | ZmPG30 & ZmPG28 | Tandem | 0.027 | 0.25 | 0.11 | 7.574 |
| 12 | ZmPG31 & ZmPG28 | Tandem | 0.03 | 0.238 | 0.125 | 7.222 |
| 13 | ZmPG33 & ZmPG32 | Tandem | 0.05 | 0.346 | 0.143 | 10.499 |
| 14 | ZmPG32 & ZmPG28 | Tandem | 0.11 | 0.608 | 0.181 | 18.424 |
| 15 | ZmPG35 & ZmPG29 | Tandem | 0.03 | 0.468 | 0.065 | 14.188 |
| 16 | ZmPG31 & ZmPG35 | Tandem | 0.033 | 0.445 | 0.073 | 13.474 |
| 17 | ZmPG30 & ZmPG35 | Tandem | 0.032 | 0.463 | 0.07 | 14.027 |
| 18 | ZmPG36 & ZmPG29 | Tandem | 0.035 | 0.469 | 0.074 | 14.2 |
| 19 | ZmPG31 & ZmPG36 | Tandem | 0.037 | 0.448 | 0.083 | 13.572 |
| 20 | ZmPG30 & ZmPG36 | Tandem | 0.037 | 0.466 | 0.079 | 14.106 |
| 21 | ZmPG32 & ZmPG35 | Tandem | 0.043 | 0.487 | 0.089 | 14.766 |
| 22 | ZmPG35 & ZmPG28 | Tandem | 0.046 | 0.488 | 0.095 | 14.774 |
| 23 | ZmPG32 & ZmPG36 | Tandem | 0.05 | 0.536 | 0.092 | 16.242 |
| 24 | ZmPG36 & ZmPG28 | Tandem | 0.051 | 0.545 | 0.093 | 16.523 |
| 25 | ZmPG21 & ZmPG15 | Segmental | 0.044 | 0.615 | 0.071 | 18.624 |
| 26 | ZmPG43 & ZmPG8 | Segmental | 0.042 | 0.137 | 0.307 | 4.151 |
| 27 | ZmPG47 & ZmPG8 | Segmental | 1.001 | 0.998 | 1.003 | 30.237 |
| 28 | ZmPG51 & ZmPG11 | Segmental | 0.935 | 1.357 | 0.689 | 41.115 |
| 29 | ZmPG49 & ZmPG12 | Segmental | 0.705 | 1.872 | 0.377 | 56.732 |
| 30 | ZmPG42 & ZmPG1 | Segmental | 0.165 | 1.385 | 0.119 | 41.982 |
| 31 | ZmPG23 & ZmPG4 | Segmental | 0.934 | 1.342 | 0.696 | 40.666 |
| 32 | ZmPG47 & ZmPG43 | Segmental | 0.649 | 0.246 | 2.634 | 7.464 |
| 33 | ZmPG46 & ZmPG44 | Segmental | 1.078 | 0.842 | 1.281 | 25.505 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Hou, Q.; Wang, L.; Zhang, T.; Zhao, W.; Yan, T.; Zhao, L.; Li, J.; Wan, X. Genome-Wide Identification and Characterization of Polygalacturonase Gene Family in Maize (Zea mays L.). Int. J. Mol. Sci. 2021, 22, 10722. https://doi.org/10.3390/ijms221910722
Lu L, Hou Q, Wang L, Zhang T, Zhao W, Yan T, Zhao L, Li J, Wan X. Genome-Wide Identification and Characterization of Polygalacturonase Gene Family in Maize (Zea mays L.). International Journal of Molecular Sciences. 2021; 22(19):10722. https://doi.org/10.3390/ijms221910722
Chicago/Turabian StyleLu, Lu, Quancan Hou, Linlin Wang, Tianye Zhang, Wei Zhao, Tingwei Yan, Lina Zhao, Jinping Li, and Xiangyuan Wan. 2021. "Genome-Wide Identification and Characterization of Polygalacturonase Gene Family in Maize (Zea mays L.)" International Journal of Molecular Sciences 22, no. 19: 10722. https://doi.org/10.3390/ijms221910722






