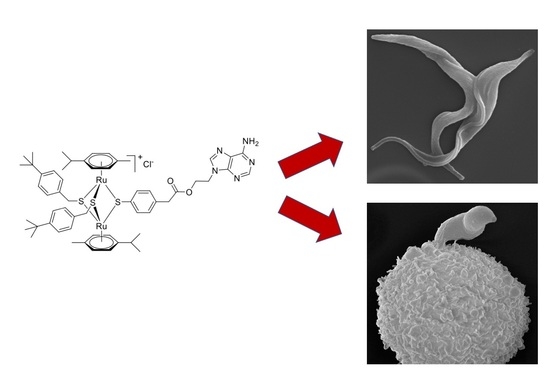
Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma gondii and Trypanosoma brucei
Abstract
:1. Introduction
2. Results
2.1. Compound 1 Inhibits Parasite Proliferation and Targets the Parasite Mitochondrion
2.2. Identification of Compound-Binding Proteins in T. gondii and T. brucei
2.3. The Major Compound 1-Binding Proteins
2.4. Compound 1 Impairs the Stability of the T. brucei ATP-Synthase Complex
3. Discussion
4. Materials and Methods
4.1. Culture Media, Biochemicals and Compounds
4.2. In Vitro Culture of Parasites
4.3. In Vitro Proliferation/Viability Measurements
4.4. Transmission Electron Microscopy
4.5. Protein Extraction and Affinity Chromatography
4.6. Proteomic Analysis of the Eluted Proteins by Mass Spectrometry
4.7. Crude Membrane Fractions and Native Polyacrylamide Gel Electrophoresis (PAGE)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adl, S.M.; Simpson, A.G.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matta, S.K.; Rinkenberger, N.; Dunay, I.R.; Sibley, L.D. Toxoplasma gondii infection and its implications within the central nervous system. Nat. Rev. Microbiol. 2021, 19, 467–480. [Google Scholar] [CrossRef]
- Hampton, M.M. Congenital toxoplasmosis: A review. Neonatal Netw. 2015, 34, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Dunay, I.R.; Gajurel, K.; Dhakal, R.; Liesenfeld, O.; Montoya, J.G. Treatment of toxoplasmosis: Historical perspective, animal models, and current clinical practice. Clin. Microbiol. Rev. 2018, 31, e00057-17. [Google Scholar] [CrossRef] [Green Version]
- Franco, J.R.; Simarro, P.P.; Diarra, A.; Ruiz-Postigo, J.A.; Jannin, J.G. The journey towards elimination of gambiense human African trypanosomiasis: Not far, nor easy. Parasitology 2014, 141, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; HP, D.E.K.; Barrett, M.P. The animal trypanosomiases and their chemotherapy: A review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef] [PubMed]
- Buscher, P.; Cecchi, G.; Jamonneau, V.; Priotto, G. Human African trypanosomiasis. Lancet 2017, 390, 2397–2409. [Google Scholar] [CrossRef]
- Dubey, J.P. Toxoplasmosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, NJ, USA, 2010; pp. 313–314. [Google Scholar]
- Attias, M.; Teixeira, D.E.; Benchimol, M.; Vommaro, R.C.; Crepaldi, P.H.; De Souza, W. The life-cycle of Toxoplasma gondii reviewed using animations. Parasites Vectors 2020, 13, 588. [Google Scholar] [CrossRef]
- Romero-Meza, G.; Mugnier, M.R. Trypanosoma brucei. Trends Parasitol. 2020, 36, 571–572. [Google Scholar] [CrossRef]
- Matthews, K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005, 118, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Lujan, H.D. Trying to get rid of protozoan parasites. Curr. Opin. Microbiol. 2012, 15, 447–448. [Google Scholar] [CrossRef]
- Müller, J.; Hemphill, A. New approaches for the identification of drug targets in protozoan parasites. Int. Rev. Cell Mol. Biol 2013, 301, 359–401. [Google Scholar] [CrossRef] [PubMed]
- Basto, A.P.; Müller, J.; Rubbiani, R.; Stibal, D.; Giannini, F.; Suss-Fink, G.; Balmer, V.; Hemphill, A.; Gasser, G.; Furrer, J. Characterization of the activities of dinuclear thiolato-bridged arene ruthenium complexes against Toxoplasma gondii. Antimicrob. Agents Chemother. 2017, 61, e01031-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studer, V.; Anghel, N.; Desiatkina, O.; Felder, T.; Boubaker, G.; Amdouni, Y.; Ramseier, J.; Hungerbuhler, M.; Kempf, C.; Heverhagen, J.T.; et al. Conjugates containing two and three trithiolato-bridged dinuclear ruthenium(II)-arene units as in vitro antiparasitic and anticancer agents. Pharmaceuticals 2020, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Desiatkina, O.; Paunescu, E.; Mosching, M.; Anghel, N.; Boubaker, G.; Amdouni, Y.; Hemphill, A.; Furrer, J. Coumarin-tagged dinuclear trithiolato-bridged ruthenium(II)arene complexes: Photophysical properties and antiparasitic activity. Chembiochem 2020, 21, 2818–2835. [Google Scholar] [CrossRef] [PubMed]
- Barna, F.; Debache, K.; Vock, C.A.; Kuster, T.; Hemphill, A. In vitro effects of novel ruthenium complexes in Neospora caninum and Toxoplasma gondii tachyzoites. Antimicrob. Agents Chemother. 2013, 57, 5747–5754. [Google Scholar] [CrossRef] [Green Version]
- Basto, A.P.; Anghel, N.; Rubbiani, R.; Müller, J.; Stibal, D.; Giannini, F.; Suss-Fink, G.; Balmer, V.; Gasser, G.; Furrer, J.; et al. Targeting of the mitochondrion by dinuclear thiolato-bridged arene ruthenium complexes in cancer cells and in the apicomplexan parasite Neospora caninum. Metallomics 2019, 11, 462–474. [Google Scholar] [CrossRef] [Green Version]
- Jelk, J.; Balmer, V.; Stibal, D.; Giannini, F.; Suss-Fink, G.; Butikofer, P.; Furrer, J.; Hemphill, A. Anti-parasitic dinuclear thiolato-bridged arene ruthenium complexes alter the mitochondrial ultrastructure and membrane potential in Trypanosoma brucei bloodstream forms. Exp. Parasitol. 2019, 205, 107753. [Google Scholar] [CrossRef]
- Fernandez, M.; Arce, E.R.; Sarniguet, C.; Morais, T.S.; Tomaz, A.I.; Azar, C.O.; Figueroa, R.; Diego Maya, J.; Medeiros, A.; Comini, M.; et al. Novel ruthenium(II) cyclopentadienyl thiosemicarbazone compounds with antiproliferative activity on pathogenic trypanosomatid parasites. J. Inorg. Biochem. 2015, 153, 306–314. [Google Scholar] [CrossRef] [Green Version]
- Gazanion, E.; Vergnes, B. Protozoan Parasite Auxotrophies and Metabolic Dependencies. Exp. Suppl. 2018, 109, 351–375. [Google Scholar] [CrossRef]
- Coppens, I. Exploitation of auxotrophies and metabolic defects in Toxoplasma as therapeutic approaches. Int. J. Parasitol. 2014, 44, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Kokina, A.; Ozolina, Z.; Liepins, J. Purine auxotrophy: Possible applications beyond genetic marker. Yeast 2019, 36, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S. Three classes of C2H2 zinc finger proteins. Cell Mol. Life Sci. 2001, 58, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Wang, Q.; Guan, Z.; Wu, Y.; Shen, C.; Hong, S.; Cao, J.; Zhang, X.; Yan, C.; Yin, P. Cryo-EM structure of the human mitochondrial translocase TIM22 complex. Cell Res. 2021, 31, 369–372. [Google Scholar] [CrossRef]
- Heron, P.W.; Sygusch, J. Isomer activation controls stereospecificity of class I fructose-1,6-bisphosphate aldolases. J. Biol. Chem. 2017, 292, 19849–19860. [Google Scholar] [CrossRef] [Green Version]
- Gahura, O.; Hierro-Yap, C.; Zikova, A. Redesigned and reversed: Architectural and functional oddities of the trypanosomal ATP synthase. Parasitology 2021, 148, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, J.J.; King, M.S.; Zogg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R.S. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 2019, 176, 435–447.e15. [Google Scholar] [CrossRef] [Green Version]
- Luevano-Martinez, L.A.; Girard, R.; Alencar, M.B.; Silber, A.M. ATP regulates the activity of an alternative oxidase in Trypanosoma brucei. FEBS Lett. 2020, 594, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Verner, Z.; Basu, S.; Benz, C.; Dixit, S.; Dobakova, E.; Faktorova, D.; Hashimi, H.; Horakova, E.; Huang, Z.; Paris, Z.; et al. Malleable mitochondrion of Trypanosoma brucei. Int. Rev. Cell Mol. Biol. 2015, 315, 73–151. [Google Scholar] [CrossRef]
- Kang, Y.; Fielden, L.F.; Stojanovski, D. Mitochondrial protein transport in health and disease. Semin. Cell Dev. Biol 2018, 76, 142–153. [Google Scholar] [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Winzer, P.; Müller, J.; Aguado-Martinez, A.; Rahman, M.; Balmer, V.; Manser, V.; Ortega-Mora, L.M.; Ojo, K.K.; Fan, E.; Maly, D.J.; et al. In vitro and in vivo effects of the bumped kinase inhibitor 1294 in the related cyst-forming apicomplexans Toxoplasma gondii and Neospora caninum. Antimicrob. Agents Chemother. 2015, 59, 6361–6374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, S.; Müller, J.; Suana, A.; Hemphill, A. Vaccination with microneme protein NcMIC4 increases mortality in mice inoculated with Neospora caninum. J. Parasitol. 2007, 93, 1046–1055. [Google Scholar] [CrossRef]
- Müller, J.; Hemphill, A. Identification of a host cell target for the thiazolide class of broad-spectrum anti-parasitic drugs. Exp. Parasitol. 2011, 128, 145–150. [Google Scholar] [CrossRef]
- Anghel, N.; Balmer, V.; Müller, J.; Winzer, P.; Aguado-Martinez, A.; Roozbehani, M.; Pou, S.; Nilsen, A.; Riscoe, M.; Doggett, J.S.; et al. Endochin-like quinolones exhibit promising efficacy against Neospora caninum in vitro and in experimentally infected pregnant mice. Front. Vet. Sci 2018, 5, 285. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.; Heller, M.; Uldry, A.C.; Braga, S.; Müller, N. Nitroreductase activites in Giardia lamblia: ORF 17150 encodes a quinone reductase with nitroreductase activity. Pathogens 2021, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charrière, F.; Helgadottir, S.; Horn, E.K.; Soll, D.; Schneider, A. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2006, 103, 6847–6852. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]









| Compound | MW | IC50 T. gondii (nM) * | IC50 T. brucei (nM) |
|---|---|---|---|
| 1 | 1193 | 59 | 29 |
| 2 | 1032 | >1000 | >1000 |
| 3 | 179 | >1000 | >1000 |
| ORF Number | Annotation | iBAQ (% of Total) |
|---|---|---|
| TGME49_319730 | YOU2 family C2C2 zinc finger protein | 30.3 ± 1.5 |
| TGME49_316710 | Hypothetical protein | 7.1 ± 1.1 |
| TGME49_278270 | Nucleolar protein, structural component of H/ACA snoRNPs, putative | 5.1 ± 1.0 |
| TGME49_215350 | Hypothetical protein | 4.6 ± 0.2 |
| TGME49_263990 | Hypothetical protein | 4.3 ± 0.3 |
| TGME49_214940 | MIC2-associated protein M2AP | 3.8 ± 0.3 |
| TGME49_221510 | Hypothetical protein | 2.6 ± 0.3 |
| TGME49_236040 | Fructose-1,6-bisphosphate aldolase | 2.5 ± 0.3 |
| TGME49_221620 | Beta-tubulin, putative | 1.9 ± 0.5 |
| TGME49_264040 | Hypothetical protein | 1.9 ± 0.1 |
| TGME49_288245 | Hypothetical protein | 1.8 ± 0.1 |
| TGME49_215430 | Hypothetical protein | 1.6 ± 0.3 |
| TGME49_294670 | Translation initiation factor 3 subunit | 1.5 ± 0.1 |
| TGME49_226410 | EF-1 guanine nucleotide exchange domain-containing protein | 1.3 ± 0.3 |
| TGME49_218410 | Ribosomal protein RPP0 | 1.1 ± 0.2 |
| TGME49_245620 | Ribosomal-ubiquitin protein RPS27A | 1.0 ± 0.2 |
| TGME49_247770 | Hypothetical protein | 0.9 ± 0.1 |
| TGME49_289830 | Eukaryotic initiation factor-3, delta subunit, putative | 0.9 ± 0.1 |
| TGME49_287210 | Proteasome subunit alpha2, protease of the acylase family | 0.8 ± 0.1 |
| TGME49_203630 | Ribosomal protein RPL44 | 0.8 ± 0.1 |
| ORF Number | Annotation | iBAQ (% of Total) |
|---|---|---|
| Tb927.5.3090 | Mitochondrial ATP synthase subunit, putative | 24.9 ± 4.6 |
| Tb927.10.5050 | Mitochondrial ATP synthase epsilon chain | 20.0 ± 4.2 |
| Tb927.5.2930 | Mitochondrial ATP synthase subunit, putative | 13.3 ± 1.5 |
| Tb927.5.1160 | Degradation arginine-rich protein for mis-folding, putative | 4.1 ± 0.6 |
| Tb927.11.600 | Mitochondrial ATP synthase subunit, putative | 3.4 ± 0.5 |
| Tb927.3.3330 | Heat shock protein 20, putative | 2.4 ± 0.2 |
| Tb927.4.4910.1 | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor, putative | 2.3 ± 0.3 |
| Tb927.3.2880 | Mitochondrial ATP synthase subunit, putative | 2.3 ± 0.3 |
| Tb927.3.3750 | Paraflagellar rod component, putative | 2.0 ± 0.0 |
| Tb927.10.13110 | Outer arm dynein light chain 7 | 2.0 ± 0.6 |
| Tb927.10.4310 | Prohibitin 2, putative | 1.6 ± 0.1 |
| Tb927.4.3590 | Translation elongation factor 1-beta, putative | 1.6 ± 0.3 |
| Tb927.7.2780 | Hypothetical protein, conserved | 1.5 ± 0.7 |
| Tb927.6.4250 | Hypothetical protein, conserved | 1.4 ± 0.2 |
| Tb927.9.4680 | Eukaryotic initiation factor 4A-1 | 1.4 ± 0.1 |
| Tb927.6.3290 | Intraflagellar transport protein 20 | 1.2 ± 0.3 |
| Tb927.8.4810 | Prohibitin 1 | 1.1 ± 0.1 |
| Tb927.10.2080 | Hypothetical protein, conserved | 1.1 ± 0.4 |
| Tb927.5.4500 | Ras-like small GTPase, putative | 1.0 ± 0.1 |
| Tb927.3.5550 | Intraflagellar transport protein 27 | 1.0 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anghel, N.; Müller, J.; Serricchio, M.; Jelk, J.; Bütikofer, P.; Boubaker, G.; Imhof, D.; Ramseier, J.; Desiatkina, O.; Păunescu, E.; et al. Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma gondii and Trypanosoma brucei. Int. J. Mol. Sci. 2021, 22, 10787. https://doi.org/10.3390/ijms221910787
Anghel N, Müller J, Serricchio M, Jelk J, Bütikofer P, Boubaker G, Imhof D, Ramseier J, Desiatkina O, Păunescu E, et al. Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma gondii and Trypanosoma brucei. International Journal of Molecular Sciences. 2021; 22(19):10787. https://doi.org/10.3390/ijms221910787
Chicago/Turabian StyleAnghel, Nicoleta, Joachim Müller, Mauro Serricchio, Jennifer Jelk, Peter Bütikofer, Ghalia Boubaker, Dennis Imhof, Jessica Ramseier, Oksana Desiatkina, Emilia Păunescu, and et al. 2021. "Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma gondii and Trypanosoma brucei" International Journal of Molecular Sciences 22, no. 19: 10787. https://doi.org/10.3390/ijms221910787
APA StyleAnghel, N., Müller, J., Serricchio, M., Jelk, J., Bütikofer, P., Boubaker, G., Imhof, D., Ramseier, J., Desiatkina, O., Păunescu, E., Braga-Lagache, S., Heller, M., Furrer, J., & Hemphill, A. (2021). Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma gondii and Trypanosoma brucei. International Journal of Molecular Sciences, 22(19), 10787. https://doi.org/10.3390/ijms221910787