Transcriptome-Wide Analysis Reveals a Role for Extracellular Matrix and Integrin Receptor Genes in Otic Neurosensory Differentiation from Human iPSCs
Abstract
:1. Introduction
2. Results
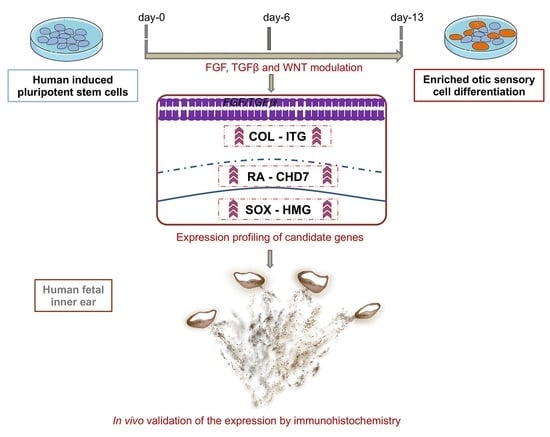
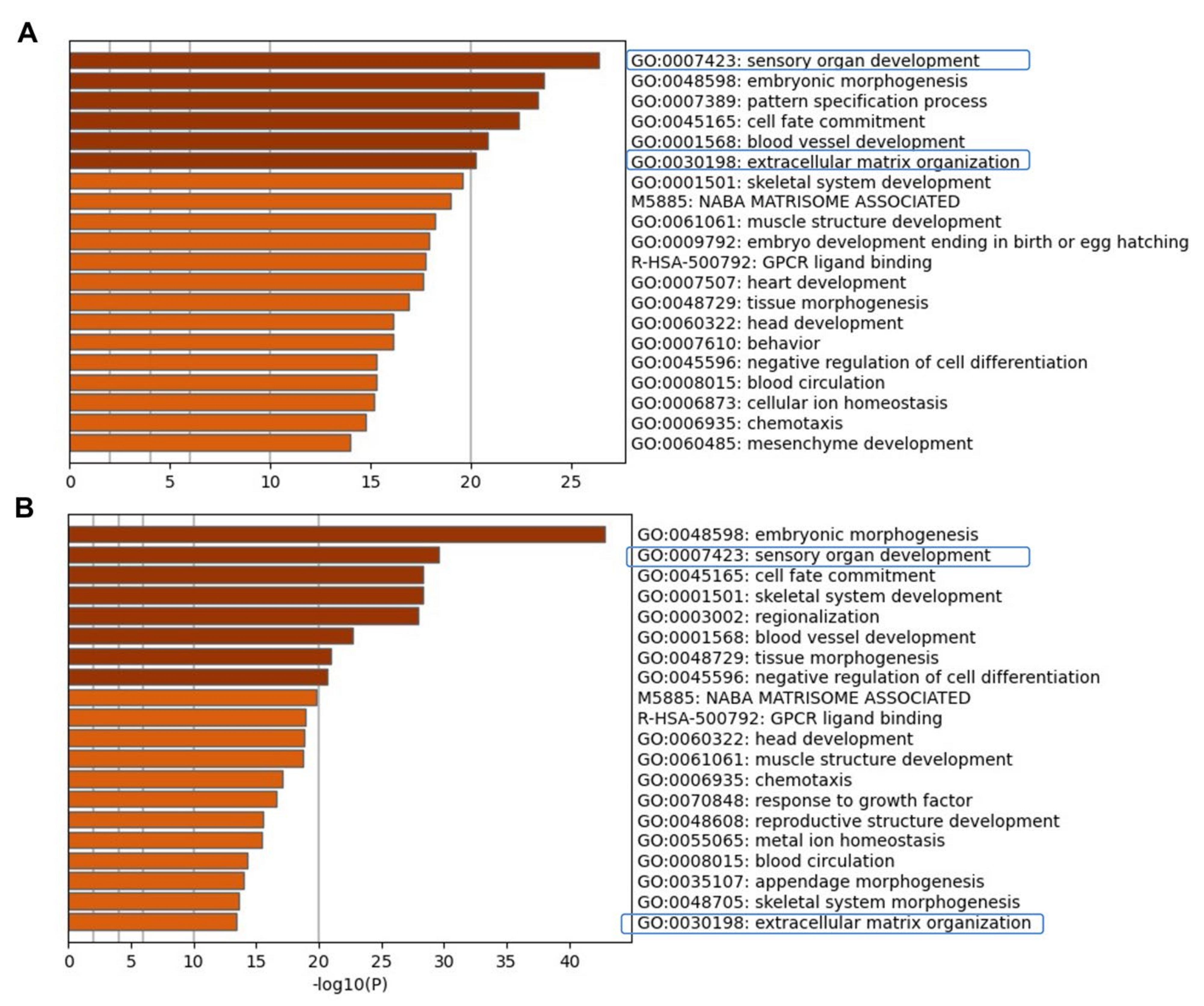
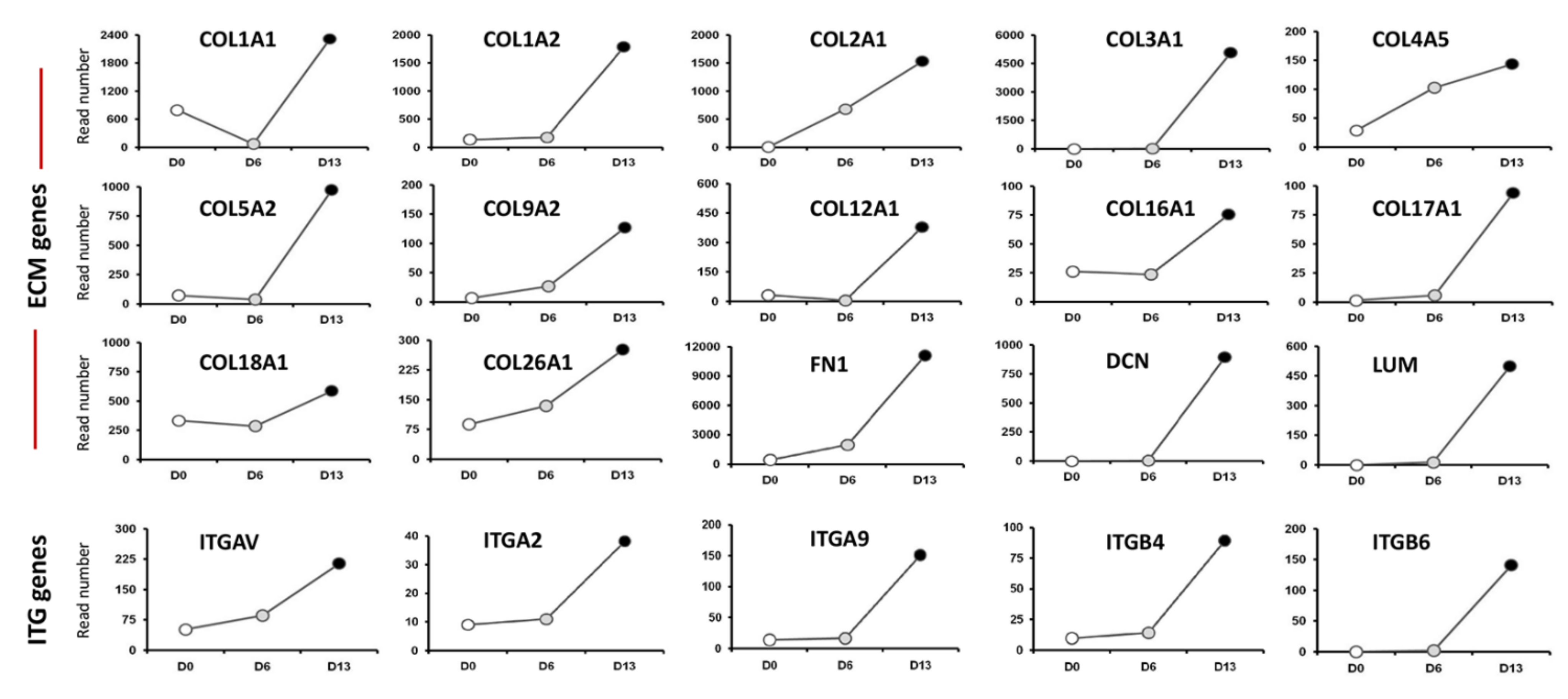
2.1. Transcriptome Analysis during hiPSC Differentiation into Otic Neurosensory Lineage
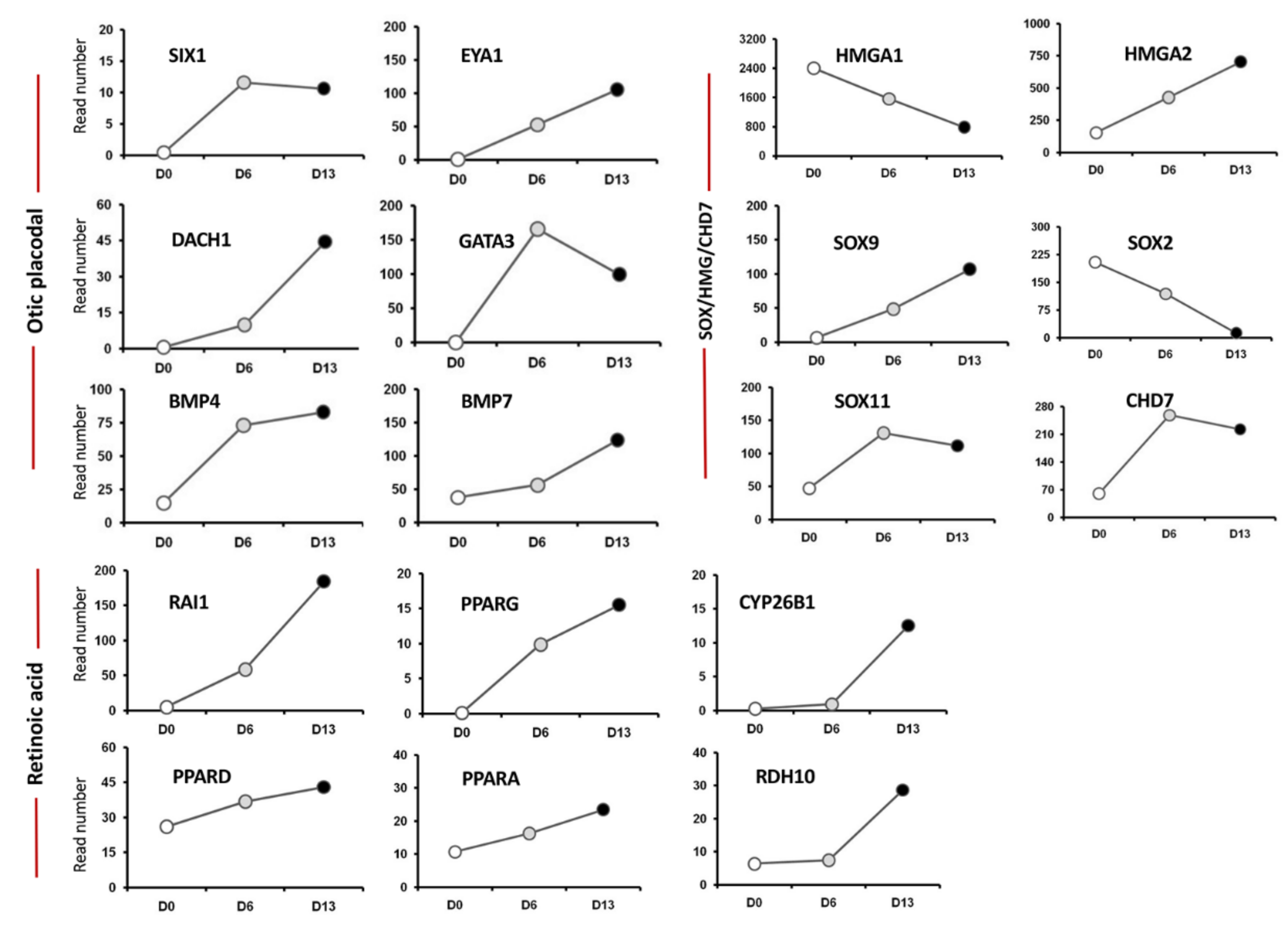
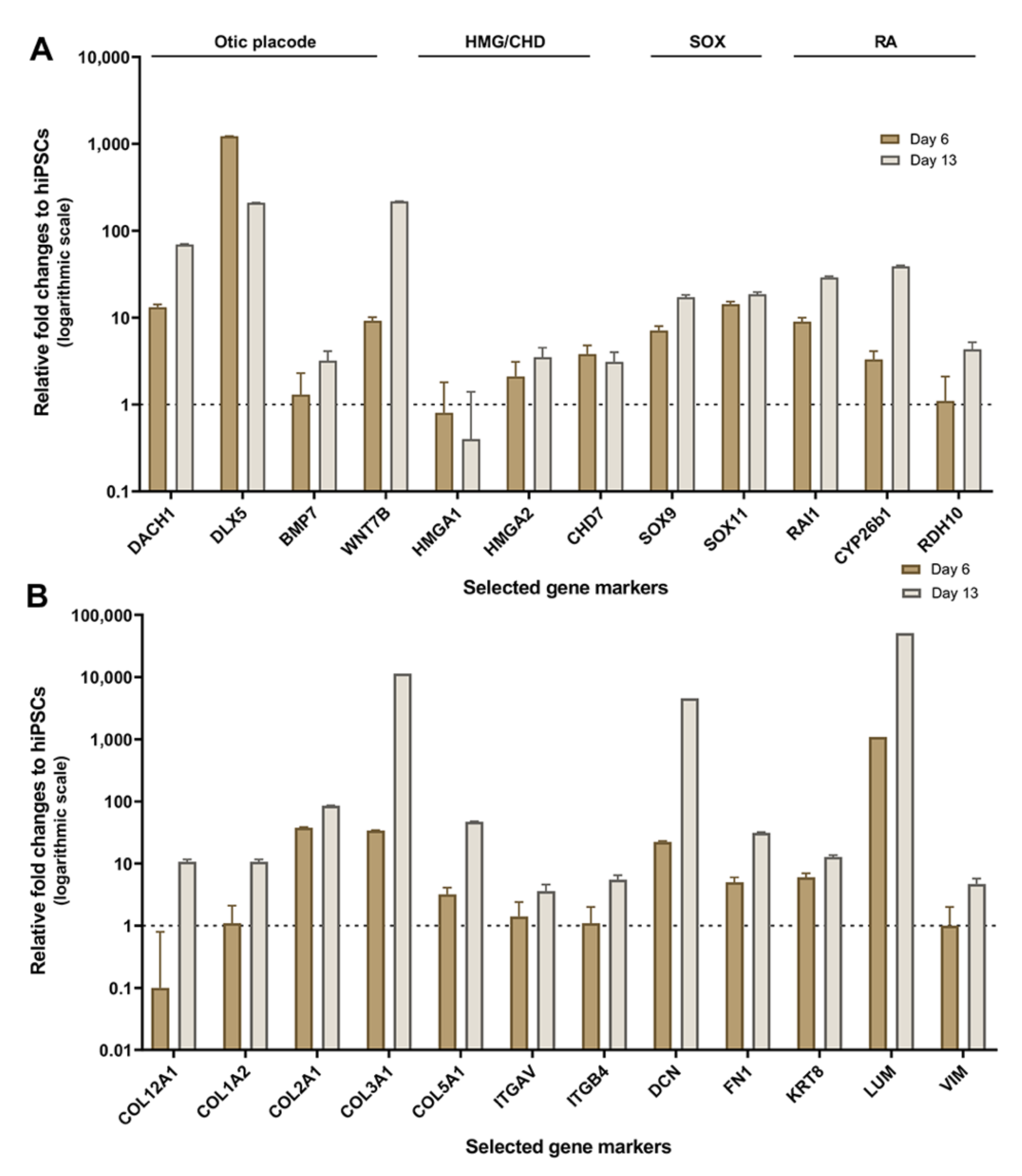
2.2. Validation of HTS Results by Quantitative PCR
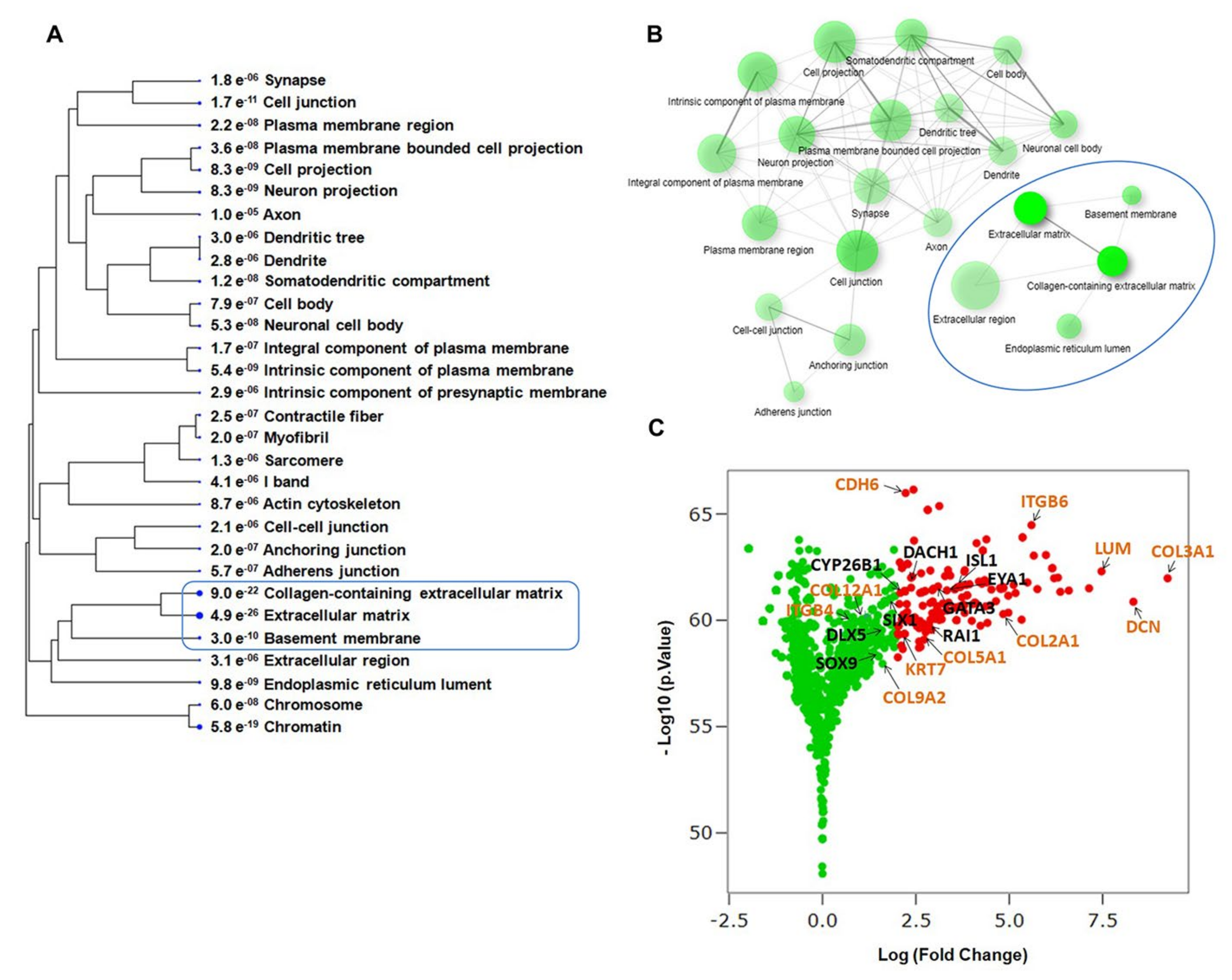
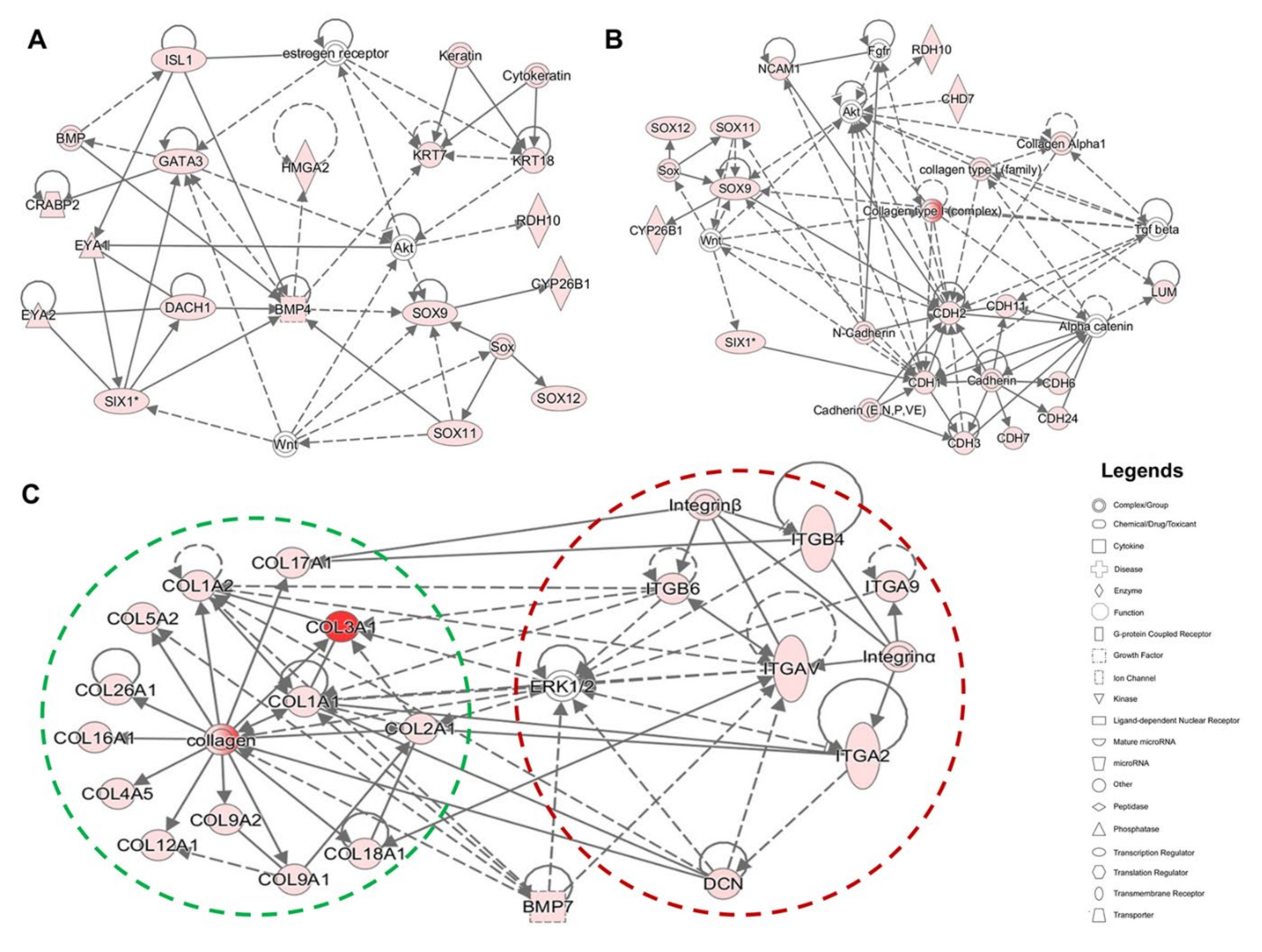
2.3. Evaluating Relationships among Differentially Expressed Genes through Network Building
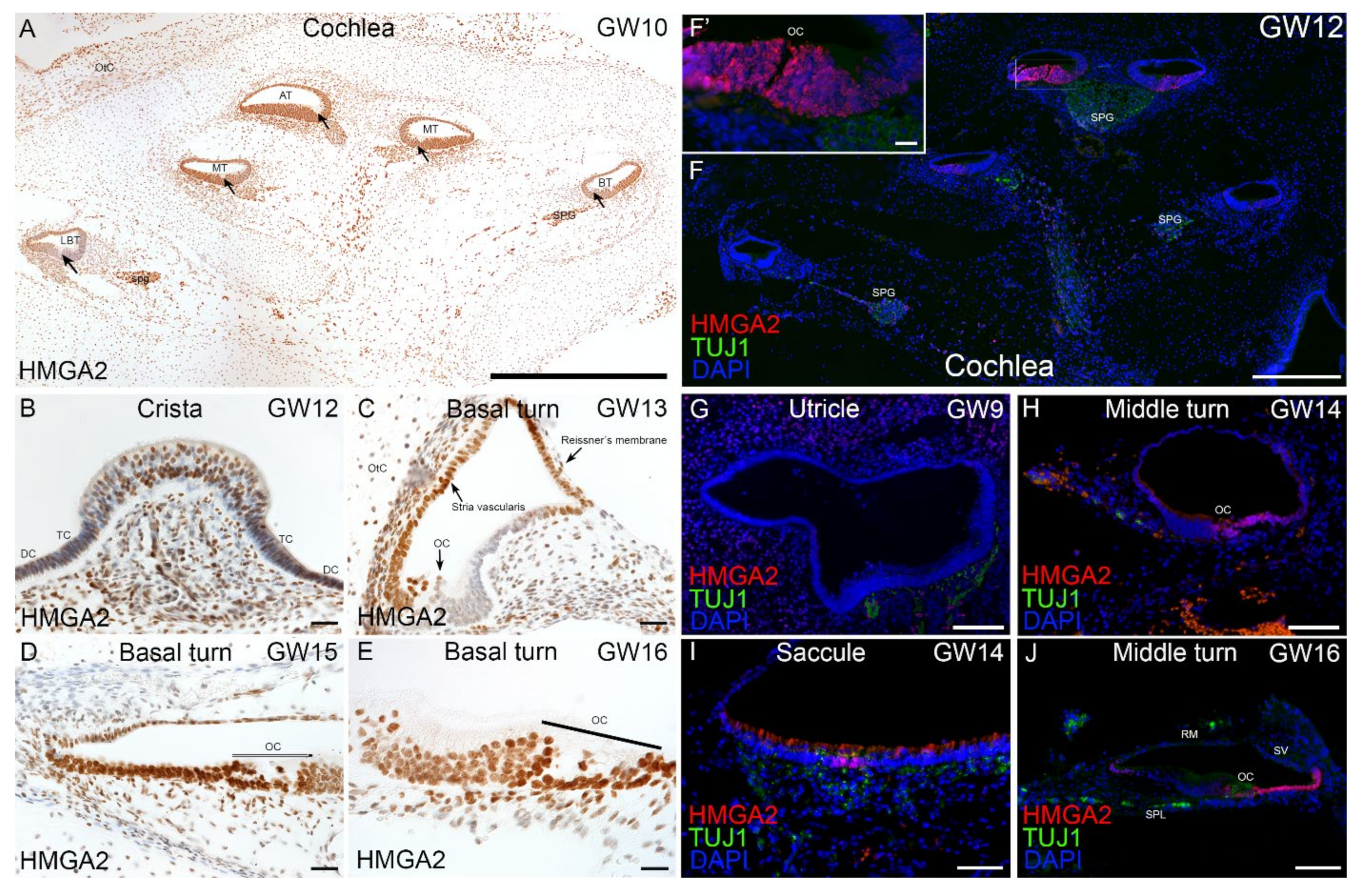
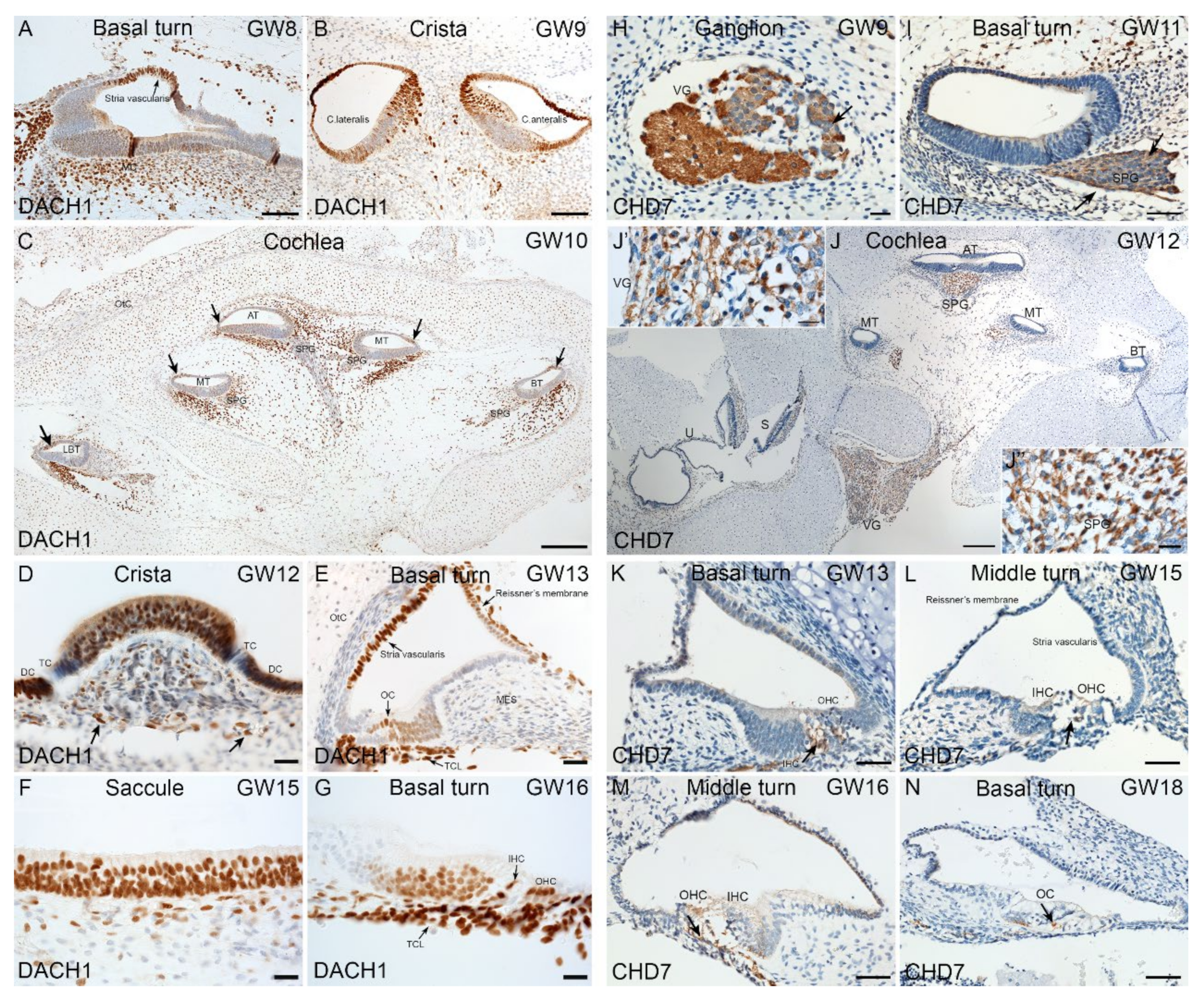
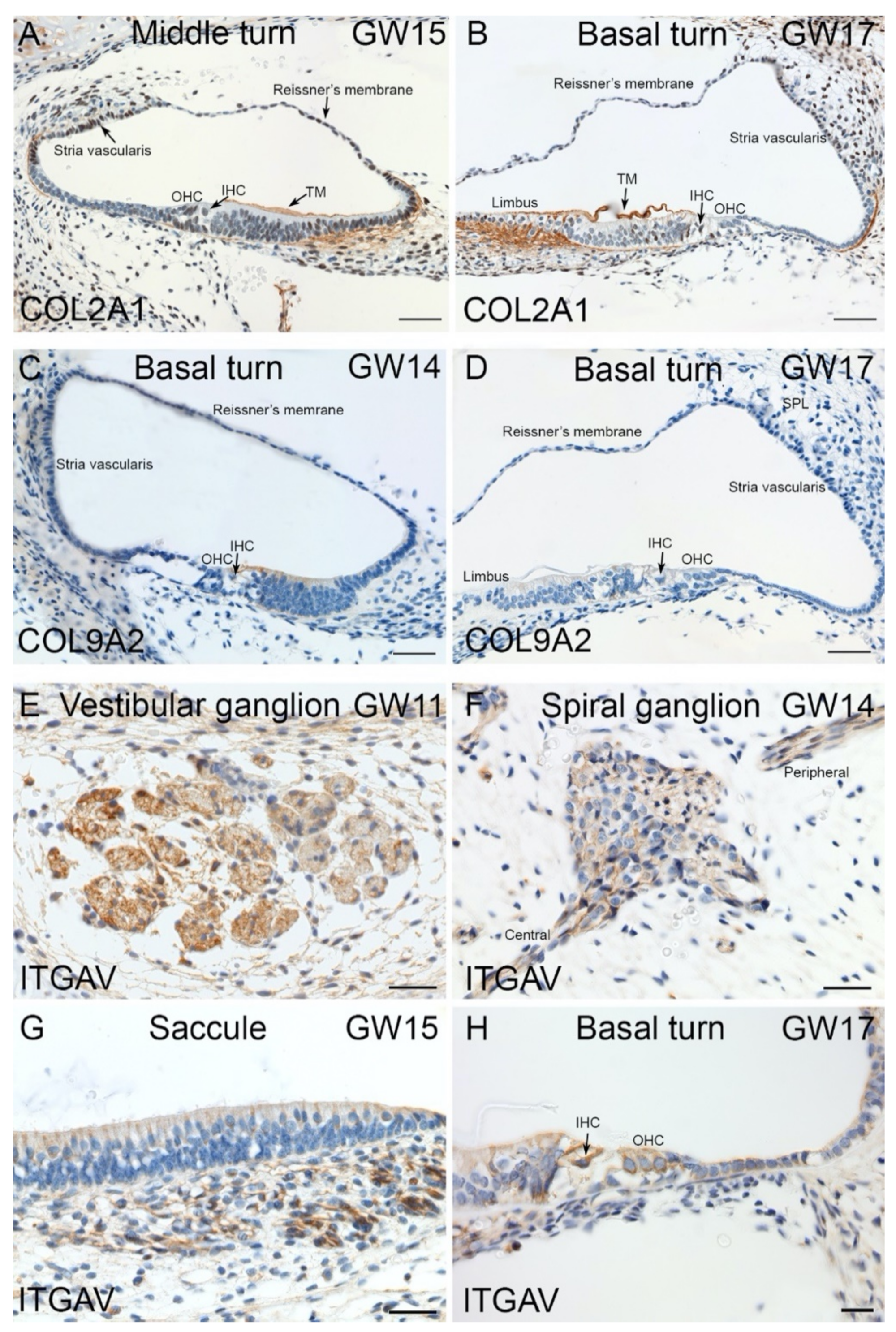
2.4. Immunohistochemical Validation of Select Markers in Human Fetal Inner Ear
3. Discussion
3.1. Enrichment of ECM and ITG during hiPSC Differentiation into Otic Sensory Lineage
3.2. Enrichment of CHD7-RA during Otic Sensory Differentiation
3.3. Enrichment of SOX/HMGA during Otic Sensory Differentiation
3.4. Expression of HL Genes during hiPSC Differentiation and in Fetal Human Inner Ear
4. Materials and Methods
4.1. Cell Line and Culture Conditions
4.2. Otic Placode Induction and Otic Sensory Differentiation
4.3. RNA-Seq Data Analysis
4.4. RNA Processing and qRT-PCR
4.5. Ethics Approval of Fetal Specimens
4.6. Tissue Preparation, Histology and Immunohistochemistry on Paraffin Sections
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wu, P.Z.; O’Malley, J.T.; de Gruttola, V.; Liberman, M.C. Age-related hearing loss is dominated by damage to inner ear sensory cells, not the cellular battery that powers them. J. Neurosci. 2020, 40, 357–6366. [Google Scholar] [CrossRef] [PubMed]
- Schilder, A.G.M.; Su, M.P.; Blackshaw, H.; Lustig, L.; Staecker, H.; Lenarz, T.; Safieddine, S.; Gomes-Santos, C.S.; Holme, R.; Warnecke, A. Hearing protection, restoration, and regeneration: An overview of emerging therapeutics for inner ear and central hearing disorders. Otol. Neurotol. 2019, 40, 559–570. [Google Scholar] [CrossRef]
- Ealy, M.; Ellwanger, D.C.; Kosaric, N.; Stapper, A.P.; Heller, S. Single-cell analysis delineates a trajectory toward the human early otic lineage. Proc. Natl. Acad. Sci. USA 2016, 113, 8508–8513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahlou, H.; Nivet, E.; Lopez-Juarez, A.; Fontbonne, A.; Assou, S.; Zine, A. Enriched differentiation of human otic sensory progenitor cells derived from induced pluripotent stem cells. Front. Mol. Neurosci. 2018, 11, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boddy, S.L.; Romero-Guevara, R.; Ji, A.-R.; Unger, C.; Corns, L.; Marcotti, W.; Rivolta, M.N. Generation of otic lineages from integration-free human-induced pluripotent stem cells reprogrammed by mRNAs. Stem Cells Int. 2020, 2020, 3692937. [Google Scholar] [CrossRef] [PubMed]
- Gunewardene, N.; Bergen, N.V.; Crombie, D.; Needham, K.; Dottori, M.; Nayagam, B.A. Directing human induced pluripotent stem cells into a neurosensory lineage for auditory neuron replacement. BioRes. Open Access 2014, 3, 162–175. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-P.; Koehler, K.R.; Mikosz, A.M.; Hashino, E.; Holt, J.R. Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat. Commun. 2016, 7, 11508. [Google Scholar] [CrossRef] [Green Version]
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.-P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017, 35, 583–589. [Google Scholar] [CrossRef] [Green Version]
- Jeong, M.; O′Reilly, M.; Kirkwood, N.K.; Al-Aama, J.; Lako, M.; Kros, C.J.; Armstrong, L. Generating inner ear organoids containing putative cochlear hair cells from human pluripotent stem cells. Cell Death Dis. 2018, 9, 922. [Google Scholar] [CrossRef] [Green Version]
- Takeda, H.; Hosoya, M.; Fujioka, M.; Saegusa, C.; Saeki, T.; Miwa, T.; Okano, H.; Minoda, R. Engraftment of human pluripotent stem cell-derived progenitors in the inner ear of prenatal mice. Sci. Rep. 2018, 8, 1941. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Juarez, A.; Lahlou, H.; Ripoll, C.; Cazals, Y.; Brezun, J.M.; Wang, Q.; Edge, A.; Zine, A. Engraftment of human stem cell-derived otic progenitors in the damaged cochlea. Mol. Ther. 2019, 27, 1101–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roccio, M.; Perny, M.; Ealy, M.; Widmer, H.R.; Heller, S.; Senn, P. Molecular characterization and prospective isolation of human fetal cochlear hair cell progenitors. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Senn, P.; Mina, A.; Volkenstein, S.; Kranebitter, V.; Oshima, K.; Heller, S. Progenitor cells from the adult human inner ear. Anat. Rec. 2019, 303, 461–470. [Google Scholar] [CrossRef]
- Singh, S.; Groves, A.K. The molecular basis of craniofacial placode development. Wiley interdisciplinary reviews. Dev. Biol. 2016, 5, 363–376. [Google Scholar] [CrossRef] [Green Version]
- Locher, H.; Frijns, J.H.M.; van Iperen, L.; de Groot, J.C.M.J.; Huisman, M.A.; Chuva de Sousa Lopes, S.M. Neurosensory development and cell fate determination in the human cochlea. Neural Dev. 2013, 8, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnedeva, K.; Hudspeth, A.J. SoxC transcription factors are essential for the development of the inner ear. Proc. Natl. Acad. Sci. USA 2015, 112, 14066–14071. [Google Scholar] [CrossRef] [Green Version]
- Parisi, S.; Piscitelli, S.; Passaro, F.; Russo, T. HMGA proteins in stemness and differentiation of embryonic and adult stem cells. Int. J. Mol. Sci. 2020, 21, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micucci, J.A.; Layman, W.S.; Hurd, E.A.; Sperry, E.D.; Frank, S.F.; Durham, M.A.; Swiderski, D.L.; Skidmore, J.M.; Scacheri, P.C.; Raphael, Y.; et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet. 2013, 23, 434–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, K.; Sandell, L.L.; Trainor, P.A.; Wu, D.K. Retinoic acid synthesis and autoregulation mediate zonal patterning of vestibular organs and inner ear morphogenesis. Development 2020, 147, dev19207. [Google Scholar] [CrossRef]
- Kondo, T.; Johnson, S.A.; Yoder, M.C.; Romand, R.; Hashino, E. Sonic hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4789–4794. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ge, Y.; Cheng, Q.; Zhang, Q.; Fang, L.; Zheng, J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget 2018, 9, 5480–5491. [Google Scholar] [CrossRef] [Green Version]
- Kelley, M.W. Cell adhesion molecules during inner ear and hair cell development, including Notch and its ligands. Curr. Top. Dev. Biol. 2003, 57, 321–356. [Google Scholar] [CrossRef] [PubMed]
- Littlewood Evans, A.; Müller, U. Stereocilia defects in the sensory hair cells of the inner ear in mice deficient in integrin α8β1. Nat. Genet. 2000, 24, 424–428. [Google Scholar] [CrossRef] [Green Version]
- Tambalo, M.; Anwar, M.; Ahmed, M.; Streit, A. Enhancer activation by FGF signalling during otic induction. Dev. Biol. 2020, 457, 69–82. [Google Scholar] [CrossRef]
- Durruthy-Durruthy, R.; Gottlieb, A.; Hartman, B.H.; Waldhaus, J.; Laske, R.D.; Altman, R.; Heller, S. Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell 2014, 157, 964–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson Chacko, L.; Sergi, C.; Eberharter, T.; Dudas, J.; Rask-Andersen, H.; Hoermann, R.; Fritsch, H.; Fischer, N.; Glueckert, R.; Schrott-Fischer, A. Early appearance of key transcription factors influence the spatiotemporal development of the human inner ear. Cell Tissue Res. 2019, 379, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidone, T.C.; Skeeters, A.V.; Oakes, P.W.; Voth, G.A. Multiscale model of integrin adhesion assembly. PLoS Comput. Biol. 2019, 15, e1007077. [Google Scholar] [CrossRef] [Green Version]
- Fietz, S.A.; Lachmann, R.; Brandl, H.; Kircher, M.; Samusik, N.; Schroder, R.; Lakshmanaperumal, N.; Henry, I.; Vogt, J.; Riehn, A.; et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. USA 2012, 109, 11836–11841. [Google Scholar] [CrossRef] [Green Version]
- Xia, M.; Chen, Y.; He, Y.; Li, H.; Li, W. Activation of the RhoA-YAP-β-catenin signaling axis promotes the expansion of inner ear progenitor cells in 3D culture. Stem Cells 2020, 38, 860–874. [Google Scholar] [CrossRef] [Green Version]
- Morton, C.C. Genetics, genomics and gene discovery in the auditory system. Hum. Mol. Genet. 2002, 11, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Hartman, B.H.; Durruthy-Durruthy, R.; Laske, R.D.; Losorelli, S.; Heller, S. Identification and characterization of mouse otic sensory lineage genes. Front. Cell. Neurosci. 2015, 9, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, T.J.; Tomoda, K. Type II collagen distribution in rodents. Laryngoscope 1988, 98, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Khetarpal, U.; Robertson, N.G.; Yoo, T.J.; Morton, C.C. Expression and localization of COL2A1 mRNA and type II collagen in human fetal cochlea. Hear. Res. 1994, 79, 59–73. [Google Scholar] [CrossRef]
- Davies, D.; Holley, M.C. Differential expression of alpha 3 and alpha 6 integrins in the developing mouse inner ear. J. Comp. Neurol. 2002, 445, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Long, K.; Moss, L.; Laursen, L.; Boulter, L.; Ffrench-Constant, C. Integrin signalling regulates the expansion of neuroepithelial progenitors and neurogenesis via Wnt7a and Decorin. Nat. Commun. 2016, 7, 10354. [Google Scholar] [CrossRef] [Green Version]
- Toyama, K.; Ozeki, M.; Hamajima, Y.; Lin, J. Expression of the integrin genes in the developing cochlea of rats. Hear. Res. 2005, 201, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Santra, M.; Reed, C.C.; Iozzo, R.V. Decorin binds to a narrow region of the epidermal growth factor (EGF) receptor, partially overlapping but distinct from the EGF-binding epitope. J. Biol. Chem. 2002, 277, 35671–35681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glueckert, R.; Rask-Andersen, H.; Sergi, C.; Schmutzhard, J.; Mueller, B.; Beckmann, F.; Rittinger, O.; Hoefsloot, L.H.; Schrott-Fischer, A.; Janecke, A.R. Histology and synchrotron radiation-based microtomography of the inner ear in a molecularly confirmed case of CHARGE syndrome. Am. J. Med. Genet. 2010, 152A, 665–673. [Google Scholar] [CrossRef]
- Feng, W.; Khan, M.A.; Bellvis, P.; Zhu, Z.; Bernhardt, O.; Herold-Mende, C.; Liu, H.-K. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 2013, 13, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Schock, E.N.; LaBonne, C. Sorting sox: Diverse roles for sox transcription factors during neural crest and craniofacial development. Front. Physiol. 2020, 11, 606889. [Google Scholar] [CrossRef]
- Urban, N.; Guillemot, F. Neurogenesis in the embryonic and adult brain: Same regulators, different roles. Front. Cell. Neurosci. 2014, 8, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savarirayan, R.; Robertson, S.P.; Bankier, A.; Rogers, J.G. Variable expression of campomelic dysplasia in a father and his 46, XY daughter. Pediatric Pathol. Mol. Med. 2003, 22, 37–46. [Google Scholar] [CrossRef]
- Dabdoub, A.; Puligilla, C.; Jones, J.M.; Fritzsch, B.; Cheah, K.S.E.; Pevny, L.H.; Kelley, M.W. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc. Natl. Acad. Sci. USA 2008, 105, 18396–18401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.N.; Kerr, C.; Cope, L.; Zambidis, E.; Liu, C.; Hillion, J.; Belton, A.; Huso, D.L.; Resar, L.M.S. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS ONE 2012, 7, e48533. [Google Scholar] [CrossRef] [Green Version]
- Li, O.; Vasudevan, D.; Davey, C.A.; Dröge, P. High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis 2006, 44, 523–529. [Google Scholar] [CrossRef]
- Smeti, I.; Watabe, I.; Savary, E.; Fontbonne, A.; Zine, A. Correction: HMGA2, the architectural transcription factor high mobility group, is expressed in the developing and mature mouse cochlea. PLoS ONE 2020, 15, e0240732. [Google Scholar] [CrossRef] [PubMed]
- Ingham, N.J.; Pearson, S.A.; Vancollie, V.E.; Rook, V.; Lewis, M.A.; Chen, J.; Buniello, A.; Martelletti, E.; Preite, L.; Lam, C.C.; et al. Mouse screen reveals multiple new genes underlying mouse and human hearing loss. PLoS Biol. 2019, 17, e3000194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson Chacko, L.; Wertjanz, D.; Sergi, C.; Dudas, J.; Fischer, N.; Eberharter, T.; Hoermann, R.; Glueckert, R.; Fritsch, H.; Rask-Andersen, H.; et al. Growth and cellular patterning during fetal human inner ear development studied by a correlative imaging approach. BMC Dev. Biol. 2019, 19, 11. [Google Scholar] [CrossRef] [Green Version]
- Zine, A.; Messat, Y.; Fritzsch, B. A human induced pluripotent stem cell-based modular platform to challenge sensorineural hearing loss. Stem Cells 2021, 39, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Booth, C.; Fillman, C.; Shapiro, M.; Blair, M.P.; Hyland, J.C.; Ala-Kokko, L. A loss of function mutation in the COL9A2 gene causes autosomal recessive Stickler syndrome. Am. J. Med. Genet. Part A 2011, 155, 1668–1672. [Google Scholar] [CrossRef]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007, 2, 3081–3089. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Sklenar, J.; Barnes, A.P.; Minnier, J. The START App: A web-based RNAseq analysis and visualization resource. Bioinformatics 2017, 33, 447–449. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson Chacko, L.; Lahlou, H.; Steinacher, C.; Assou, S.; Messat, Y.; Dudás, J.; Edge, A.; Crespo, B.; Crosier, M.; Sergi, C.; et al. Transcriptome-Wide Analysis Reveals a Role for Extracellular Matrix and Integrin Receptor Genes in Otic Neurosensory Differentiation from Human iPSCs. Int. J. Mol. Sci. 2021, 22, 10849. https://doi.org/10.3390/ijms221910849
Johnson Chacko L, Lahlou H, Steinacher C, Assou S, Messat Y, Dudás J, Edge A, Crespo B, Crosier M, Sergi C, et al. Transcriptome-Wide Analysis Reveals a Role for Extracellular Matrix and Integrin Receptor Genes in Otic Neurosensory Differentiation from Human iPSCs. International Journal of Molecular Sciences. 2021; 22(19):10849. https://doi.org/10.3390/ijms221910849
Chicago/Turabian StyleJohnson Chacko, Lejo, Hanae Lahlou, Claudia Steinacher, Said Assou, Yassine Messat, József Dudás, Albert Edge, Berta Crespo, Moira Crosier, Consolato Sergi, and et al. 2021. "Transcriptome-Wide Analysis Reveals a Role for Extracellular Matrix and Integrin Receptor Genes in Otic Neurosensory Differentiation from Human iPSCs" International Journal of Molecular Sciences 22, no. 19: 10849. https://doi.org/10.3390/ijms221910849
APA StyleJohnson Chacko, L., Lahlou, H., Steinacher, C., Assou, S., Messat, Y., Dudás, J., Edge, A., Crespo, B., Crosier, M., Sergi, C., Schrott-Fischer, A., & Zine, A. (2021). Transcriptome-Wide Analysis Reveals a Role for Extracellular Matrix and Integrin Receptor Genes in Otic Neurosensory Differentiation from Human iPSCs. International Journal of Molecular Sciences, 22(19), 10849. https://doi.org/10.3390/ijms221910849