Effects of Rifaximin on Luminal and Wall-Adhered Gut Commensal Microbiota in Mice
Abstract
:1. Introduction
2. Results
2.1. Effects of Rifaximin on Body Weight, Weight of Body Organs and Colonic Histology
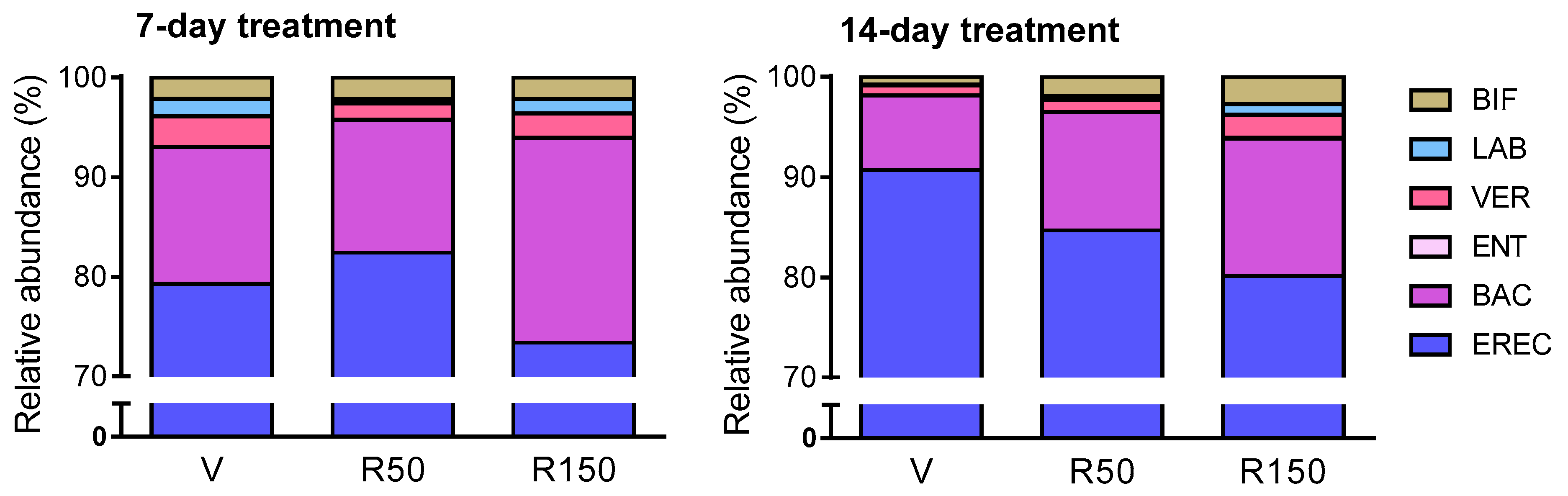
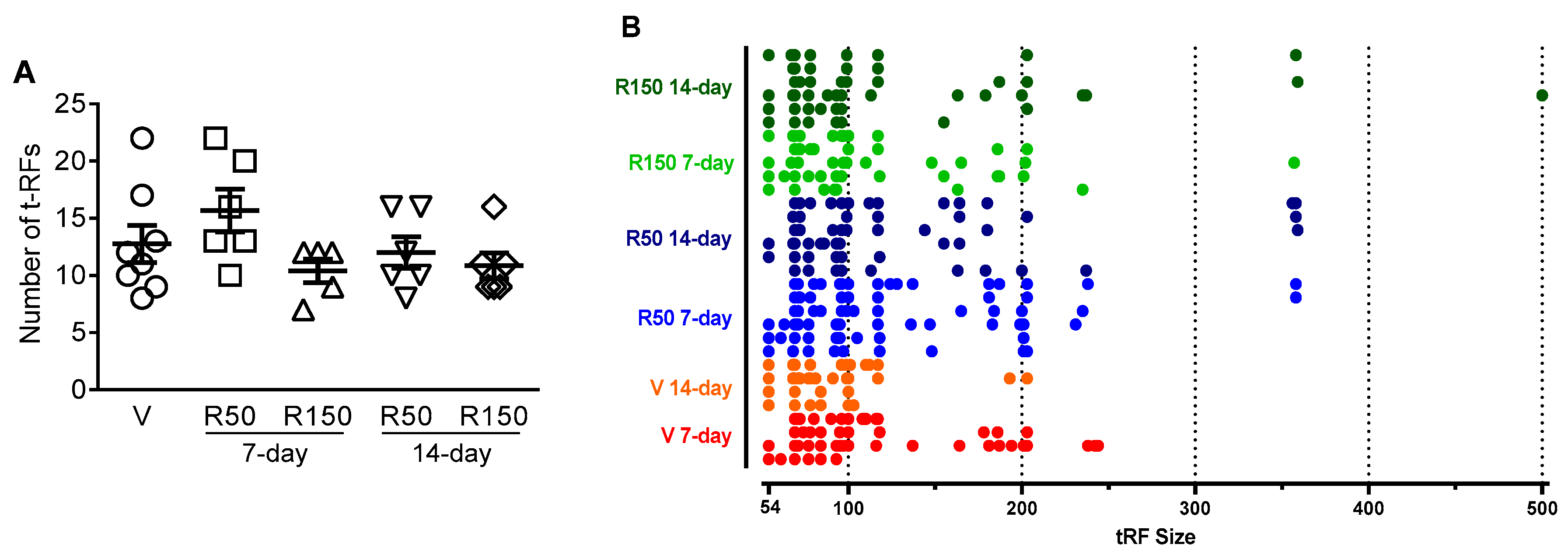
2.2. Effects of Rifaximin on Luminal GCM
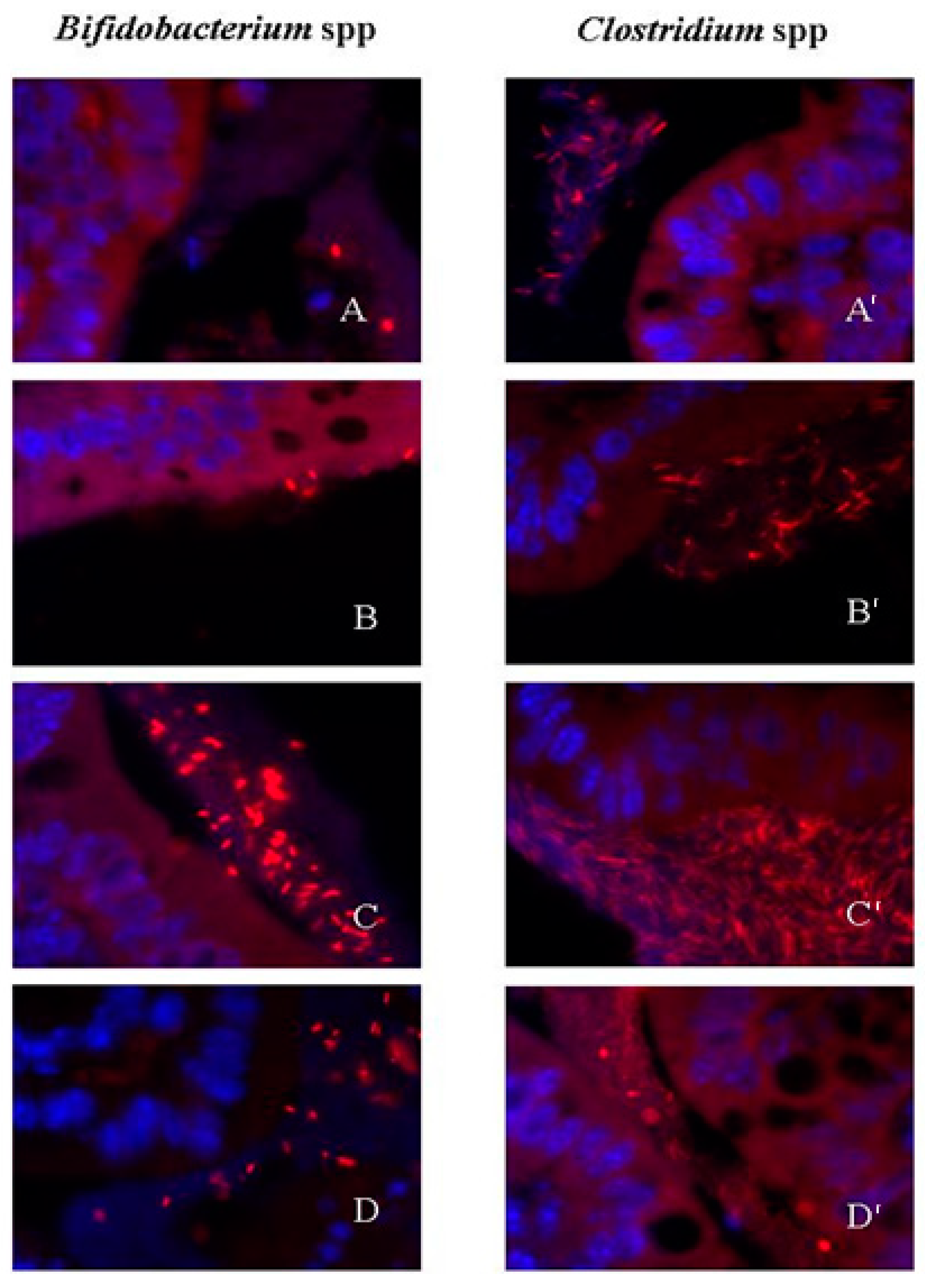
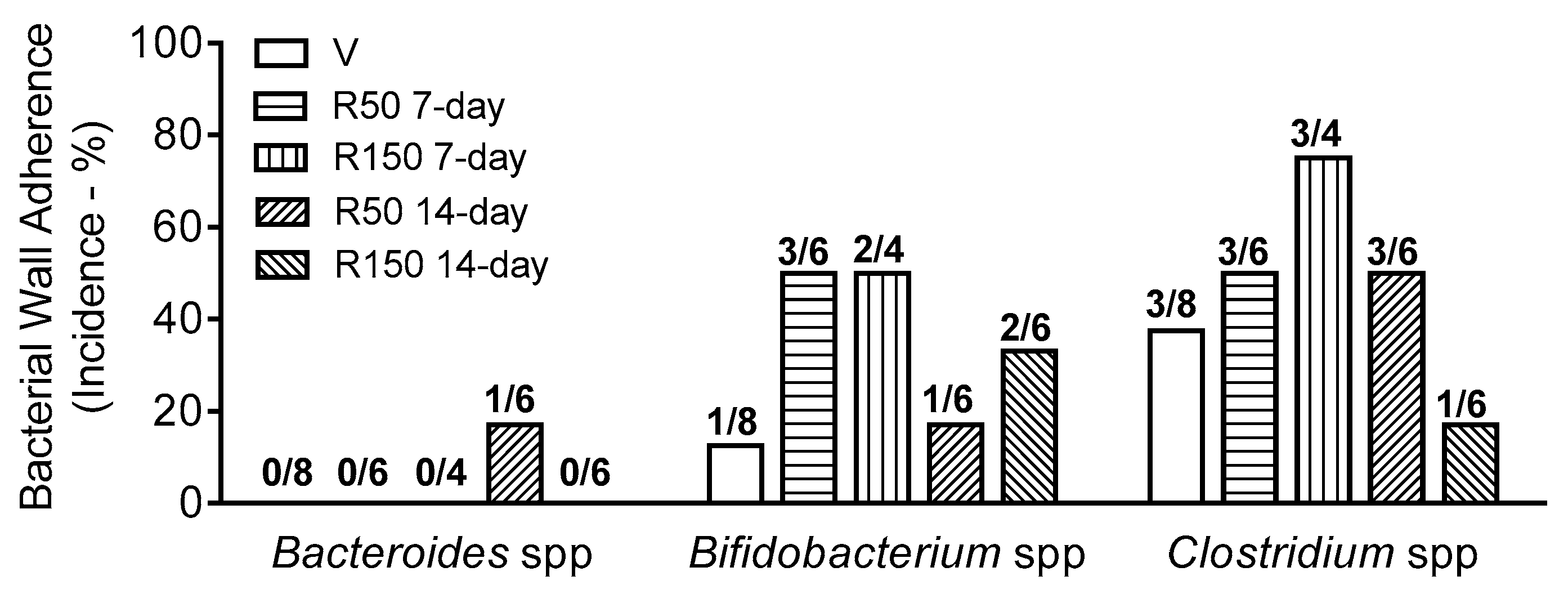
2.3. Effects of Rifaximin on Bacterial Adherence to the Colonic Wall
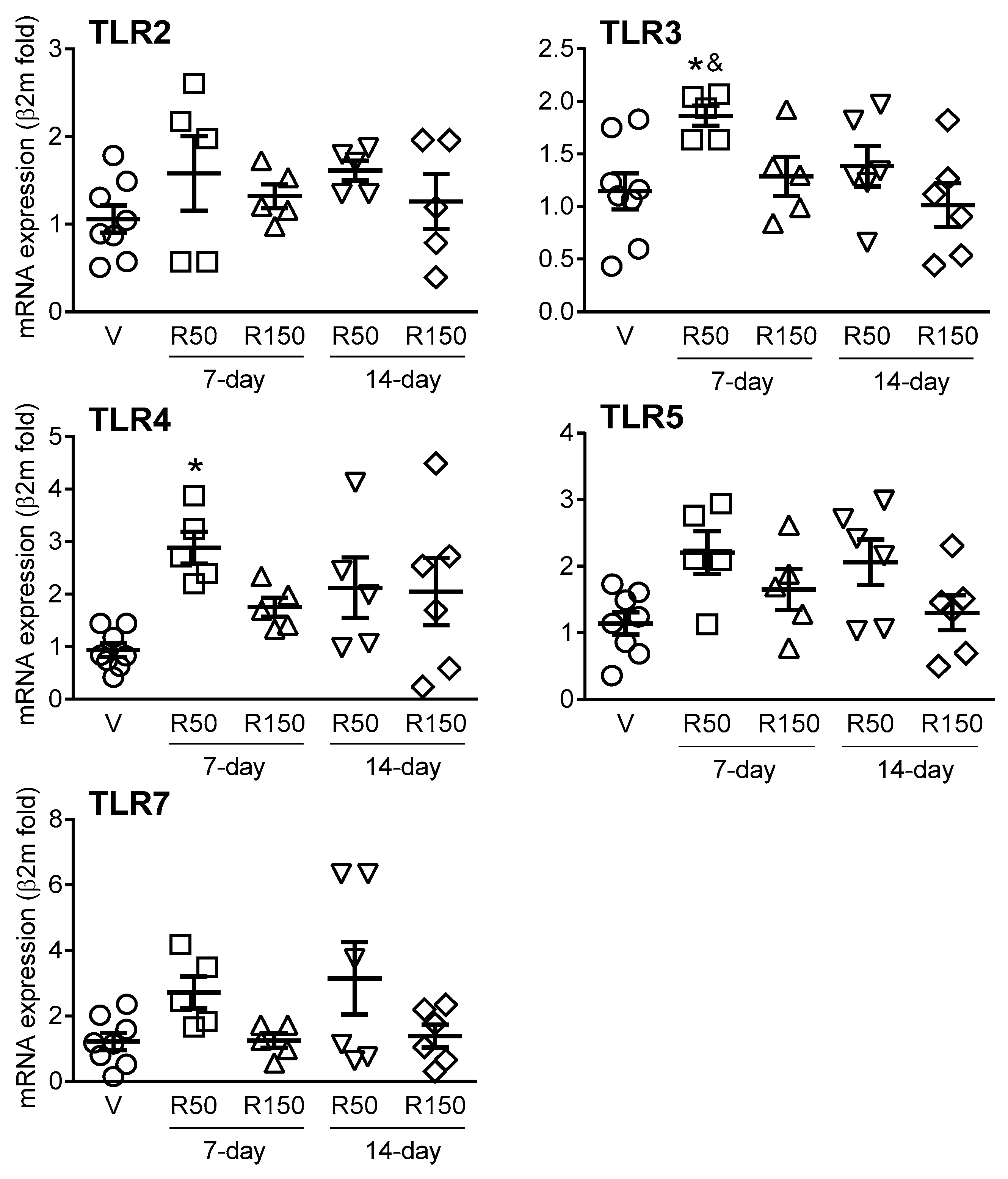
2.4. Effects of Rifaximin on Colonic Expression of TLRs and Immune-Related Markers
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Antibiotic
4.3. Experimental Protocols
4.4. Samples Collection
4.5. Histological Evaluation
4.6. Bacterial Identification by Fluorescence in Situ Hybridization (FISH)
4.7. Terminal Restriction Fragment Length Polymorphism
4.8. Colonic Expression of TLRs and Immune-Related Markers Using Quantitative Real-Time PCR (RT-qPCR)
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adachi, J.A.; DuPont, H.L. Rifaximin: A Novel Nonabsorbed Rifamycin for Gastrointestinal Disorders. Clin. Infect. Dis. 2006, 42, 541–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, S.; Vitali, B.; Klinder, A.; Kolida, S.; Ndagijimana, M.; Laghi, L.; Calanni, F.; Brigidi, P.; Gibson, G.R.; Costabile, A. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: An in vitro approach using a continuous culture colonic model system. J. Antimicrob. Chemother. 2010, 65, 2556–2565. [Google Scholar] [CrossRef] [PubMed]
- Debbia, E.A.; Maioli, E.; Roveta, S.; Marchese, A. Effects of rifaximin on bacterial virulence mechanisms at supra- and sub-inhibitory concentrations. J. Chemother. 2008, 20, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Mullen, K.D.; Sanyal, A.J.; Bass, N.M.; Poordad, F.F.; Sheikh, M.Y.; Frederick, R.T.; Bortey, E.; Forbes, W.P. Rifaximin is safe and well tolerated for long-term maintenance of remission from overt hepatic encephalopathy. Clin. Gastroenterol. Hepatol. 2014, 12, 1390–1397.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kane, J.S.; Ford, A.C. Rifaximin for the treatment of diarrhea-predominant irritable bowel syndrome. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 431–442. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Napoli, M.; Rizzatti, G.; Gasbarrini, A. The intriguing role of Rifaximin in gut barrier chronic inflammation and in the treatment of Crohn’s disease. Expert Opin. Investig. Drugs 2018, 27, 543–551. [Google Scholar] [CrossRef]
- Simrén, M.; Öhman, L. Pathogenesis of IBS: Role of inflammation, immunity and neuroimmune interactions. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 163–173. [Google Scholar]
- Raskov, H.; Burcharth, J.; Pommergaard, H.C.; Rosenberg, J. Irritable bowel syndrome, the microbiota and the gut-brain axis. Gut Microbes 2016, 7, 365–383. [Google Scholar] [CrossRef] [Green Version]
- Holtmann, G.J.; Ford, A.C.; Talley, N.J. Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol. Hepatol. 2016, 1, 133–146. [Google Scholar] [CrossRef]
- Balemans, D.; Mondelaers, S.U.; Cibert-Goton, V.; Stakenborg, N.; Aguilera-Lizarraga, J.; Dooley, J.; Liston, A.; Bulmer, D.C.; VandenBerghe, P.; Boeckxstaens, G.E.; et al. Evidence for long-term sensitization of the bowel in patients with post-infectious-IBS. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.P.; Sagrestani, G.; Delmas, E.; Wilson, K.T.; Verriere, T.G.; Dapoigny, M.; Del’homme, C.; Bernalier-Donadille, A. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Candela, M.; Maccaferri, S.; Turroni, S.; Carnevali, P.; Brigidi, P. Functional intestinal microbiome, new frontiers in prebiotic design. Int. J. Food Microbiol. 2010, 140, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Liu, Y.; Zhang, D.; Li, Y.; Xu, L. Prevalence and treatment of small intestinal bacterial overgrowth in postoperative patients with colorectal cancer. Mol. Clin. Oncol. 2016, 4, 883–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shayto, R.H.; AbouMrad, R.; Sharara, A.I. Use of rifaximin in gastrointestinal and liver diseases. World J. Gastroenterol. 2016, 22, 6638. [Google Scholar] [CrossRef]
- Bercik, P.; Verdu, E.F.; Collins, S.M. Is irritable bowel syndrome a low-grade inflammatory bowel disease? Gastroenterol. Clin. N. Am. 2005, 34, 235–245. [Google Scholar] [CrossRef]
- Cheng, J.; Shah, Y.M.; Ma, X.; Pang, X.; Tanaka, T.; Kodama, T.; Krausz, K.W.; Gonzalez, F.J. Therapeutic role of rifaximin in inflammatory bowel disease: Clinical implication of human pregnane X receptor activation. J. Pharmacol. Exp. Ther. 2010, 335, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Gao, J.; Gillilland, M., 3rd; Wu, X.; Song, I.; Kao, J.Y.; Owyang, C. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterology 2014, 146, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.L.; Xue, Q.; Jiang, Z.D.; Xu, Y.; DuPont, H.L. Pretreatment of epithelial cells with rifaximin alters bacterial attachment and internalization profiles. Antimicrob. Agents Chemother. 2010, 54, 388–396. [Google Scholar] [CrossRef] [Green Version]
- Terc, J.; Hansen, A.; Alston, L.; Hirota, S.A. Pregnane X receptor agonists enhance intestinal epithelial wound healing and repair of the intestinal barrier following the induction of experimental colitis. Eur. J. Pharm. Sci. 2014, 55, 12–19. [Google Scholar] [CrossRef]
- Sartor, R.B. Review article: The potential mechanisms of action of rifaximin in the management of inflammatory bowel diseases. Aliment. Pharmacol. Ther. 2016, 43 (Suppl. S1), 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorucci, S.; Distrutti, E.; Mencarelli, A.; Barbanti, M.; Palazzini, E.; Morelli, A. Inhibition of intestinal bacterial translocation with rifaximin modulates lamina propria monocytic cells reactivity and protects against inflammation in a rodent model of colitis. Digestion 2002, 66, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.; Vergara, P.; Martínez, V. Stress and antibiotics alter luminal and wall-adhered microbiota and enhance the local expression of visceral sensory-related systems in mice. Neurogastroenterol. Motil. 2013, 25, e515–e529. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.; Vergara, P.; Martínez, V. Environment-related adaptive changes of gut commensal microbiota do not alter colonic toll-like receptors but modulate the local expression of sensory-related systems in rats. Microb. Ecol. 2013, 66, 232–243. [Google Scholar] [CrossRef]
- Siezen, R.J.; Kleerebezem, M. The human gut microbiome: Are we our enterotypes? Microb. Biotechnol. 2011, 4, 550–553. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Brigidi, P.; Swennen, E.; Rizzello, F.; Bozzolasco, M.; Matteuzzi, D. Effects of rifaximin administration on the intestinal microbiota in patients with ulcerative colitis. J. Chemother. 2002, 14, 290–295. [Google Scholar] [CrossRef]
- DuPont, H.L.; Jiang, Z.D.; Okhuysen, P.C.; Ericsson, C.D.; de la Cabada, F.J.; Ke, S.; DuPont, M.W.; Martinez-Sandoval, F. A randomized, double-blind, placebo-controlled trial of rifaximin to prevent travelers’ diarrhea. Ann. Intern. Med. 2005, 142, 805–812. [Google Scholar] [CrossRef]
- Jin, Y.; Ren, X.; Li, G.; Li, Y.; Zhang, L.; Wang, H.; Qian, W.; Hou, X. Beneficial effects of Rifaximin in post-infectious irritable bowel syndrome mouse model beyond gut microbiota. J. Gastroenterol. Hepatol. 2018, 33, 443–452. [Google Scholar] [CrossRef]
- Miglioli, P.A.; Allerberger, F.; Calabrò, G.B.; Gaion, R.M. Effects of daily oral administration of rifaximin and neomycin on faecal aerobic flora in rats. Pharmacol. Res. 2001, 44, 373–375. [Google Scholar] [CrossRef]
- Gao, J.; Gillilland, M.G.; Owyang, C. Rifaximin, gut microbes and mucosal inflammation: Unraveling a complex relationship. Gut Microbes 2014, 5, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.; Cerdà-Cuéllar, M.; Martínez, V. Antibiotic-induced dysbiosis alters host-bacterial interactions and leads to colonic sensory and motor changes in mice. Gut Microbes 2015, 6, 10–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdú, E.F.; Bercik, P.; Verma-Gandhu, M.; Huang, X.X.; Blennerhassett, P.; Jackson, W.; Mao, Y.; Wang, L.; Rochat, F.; Collins, S.M. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 2006, 55, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, B.; Zheng, J.; Huang, J.; Zhao, Q.; Liu, J.; Su, Z.; Wang, M.; Cui, Z.; Wang, T.; et al. Rifaximin alters intestinal microbiota and prevents progression of ankylosing spondylitis in mice. Front. Cell. Infect. Microbiol. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.N.; McKenzie, R.; Durbin, A.; Carpenter, C.; Atzinger, C.B.; Haake, R.; Bourgeois, A.L. Rifaximin, a nonabsorbed oral antibiotic, prevents shigellosis after experimental challenge. Clin. Infect. Dis. 2006, 42, 1283–1288. [Google Scholar] [CrossRef] [Green Version]
- Gionchetti, P.; Rizzello, F.; Venturi, A.; Campieri, M. Probiotics in infective diarrhoea and inflammatory bowel diseases. J. Gastroenterol. Hepatol. 2000, 15, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Terán-Ventura, E.; Roca, M.; Martin, M.T.; Abarca, M.L.; Martinez, V.; Vergara, P. Characterization of housing-related spontaneous variations of gut microbiota and expression of toll-like receptors 2 and 4 in rats. Microb. Ecol. 2010, 60, 691–702. [Google Scholar] [CrossRef]
- Kang, D.J.; Kakiyama, G.; Betrapally, N.S.; Herzog, J.; Nittono, H.; Hylemon, P.B.; Zhou, H.; Carroll, I.; Yang, J.; Gillevet, P.M.; et al. Rifaximin exerts beneficial effects independent of its ability to alter microbiota composition. Clin. Transl. Gastroenterol. 2016, 7, e187. [Google Scholar] [CrossRef]
- Salzman, N.H. Microbiota–immune system interaction: An uneasy alliance. Curr. Opin. Microbiol. 2011, 14, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–143. [Google Scholar] [CrossRef]
- Kordjazy, N.; Haj-Mirzaian, A.; Haj-Mirzaian, A.; Rohani, M.M.; Gelfand, E.W.; Rezaei, N.; Abdolghaffari, A.H. Role of toll-like receptors in inflammatory bowel disease. Pharmacol. Res. 2018, 129, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Liu, S.; Zhang, Y.; Zhang, D. Toll-like receptors and inflammatory bowel disease. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frosali, S.; Pagliari, D.; Gambassi, G.; Landolfi, R.; Pandolfi, F.; Cianci, R. How the intricate interaction among Toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J. Immunol. Res. 2015, 2015, 489821. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesi, A.; García-Albiach, R.; Pozuelo, M.J.; Pintado, C.; Goñi, I.; Rotger, R. Molecular and microbiological analysis of caecal microbiota in rats fed with diets supplemented either with prebiotics or probiotics. Int. J. Food Microbiol. 2005, 98, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Sex as a biological variable in irritable bowel syndrome. Neurogastroenterol. Motil. 2020, 32, e13802. [Google Scholar] [CrossRef] [PubMed]
- Prusator, D.K.; Chang, L. Sex-related differences in GI disorders. Handb. Exp. Pharmacol. 2017, 239, 177–192. [Google Scholar]
- Terán-Ventura, E.; Aguilera, M.; Vergara, P.; Martínez, V. Specific changes of gut commensal microbiota and TLRs during indomethacin-induced acute intestinal inflammation in rats. J. Crohns Colitis 2014, 8, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Salzman, N.H.; de Jong, H.; Paterson, Y.; Harmsen, H.J.M.; Welling, G.W.; Bos, N.A. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 2002, 148, 3651–3660. [Google Scholar] [CrossRef] [Green Version]
- Selinummi, J.; Seppälä, J.; Yli-Harja, O.; Puhakka, J.A. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques 2005, 39, 859–862. [Google Scholar] [CrossRef] [Green Version]
- Højberg, O.; Canibe, N.; Poulsen, H.D.; Hedemann, M.S.; Jensen, B.B. Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets. Appl. Environ. Microbiol. 2005, 71, 2267–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitts, C.L. Terminal restriction fragment patterns: A tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2001, 2, 7–25. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Treatment Duration | Treatment | n | Body Weight at Necropsy (g) | Body Weight (% Change from Day 0) | Colon Relative Weight (mg/cm) | Cecum Relative Weight (mg/g Body Weight) |
|---|---|---|---|---|---|---|
| 7-day | Vehicle | 4 | 17.6 ± 0.6 | −4.6 ± 2.7 | 23.2 ± 1.7 | 21.5 ± 0.9 |
| Rifaximin (50 mg/kg) | 6 | 17.8 ± 0.2 | 2.7 ± 2.4 | 21.8 ± 0.8 | 25.1 ± 2.3 | |
| Rifaximin (150 mg/kg) | 5 | 18.3 ± 0.2 | −1.6 ± 1.5 | 21.3 ± 1.6 | 25.9 ± 0.9 | |
| 14-day | Vehicle | 4 | 17.4 ± 0.7 | 1.9 ± 2.2 | 21.7 ± 0.2 | 22.2 ± 0.8 |
| Rifaximin (50 mg/kg) | 6 | 18.3 ± 0.3 | −2.5 ± 2.1 | 22.9 ± 1.3 | 23.7 ± 1.9 | |
| Rifaximin (150 mg/kg) | 6 | 18.2 ± 0.2 | 2.1 ± 1.4 | 24.5 ± 0.7 | 21.2 ± 0.9 |
| Compatible Bacterial Group | tRFSize | Frequency 1 | |||||
|---|---|---|---|---|---|---|---|
| V 7-day (n = 4) | V 14-day (n = 4) | R50 7-day (n = 6) | R50 14-day (n = 6) | R150 7-day (n = 5) | R150 14-day (n = 6) | ||
| Unidentified | 54–55 | 3 (75) | 4 (100) | 4 (67) | 3 (50) | 3 (60) | 5 (83) |
| Bacillus spp./Lactococcus lactis spp. | 61–62 | 1 (25) | 2 (50) | 1 (17) | 0 (0) | 1 (20) | 1 (17) |
| Salinicoccusroseus | 63 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Burkholderia spp./Bordetella spp./Thiomonas spp./ uncultured bacterium | 67 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 1 (17) |
| Mycobacterium spp./ uncultured rumen bacterium | 68 | 0 (0) | 2 (50) | 1 (17) | 2 (33) | 1 (20) | 2 (33) |
| Uncultured rumen bacterium | 69 | 4 (100) | 4 (100) | 5 (83) | 5 (83) | 4 (80) | 6 (100) |
| Uncultured rumen bacterium/ Leptotrichia spp. | 71 | 2 (50) | 1 (25) | 4 (67) | 1 (17) | 2 (40) | 0 (0) |
| Uncultured rumen bacterium | 72 | 1 (25) | 1 (25) | 1 (17) | 3 (50) | 2 (40) | 1 (17) |
| Photorhabdus sp. | 74–75 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured bacterium | 77 | 2 (50) | 1 (25) | 3 (50) | 3 (50) | 1 (20) | 5 (83) |
| Uncultured rumen bacterium/ naphthalene-utilizing bacterium | 78 | 1 (25) | 3 (75) | 1 (17) | 1 (17) | 1 (20) | 3 (50) |
| Uncultured bacterium | 80 | 1 (25) | 0 (0) | 2 (33) | 0 (0) | 1 (20) | 0 (0) |
| Sphingomonas spp./ uncultured bacterium | 81 | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured bacterium | 84 | 3 (75) | 2 (50) | 2 (33) | 1 (17) | 0 (0) | 1 (17) |
| Desulfovibriodefluvii/ Roseiflexus spp. | 86–87 | 2 (50) | 1 (25) | 1 (17) | 2 (33) | 1 (20) | 5 (83) |
| Flavobacterium psychrophilum | 88–89 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (17) |
| Flavobacterium johnsoniae | 90 | 1 (25) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) |
| Anaeromyxobacter dehalogenans/ uncultured bacterium | 91 | 0 (0) | 1 (25) | 0 (0) | 1 (17) | 2 (40) | 0 (0) |
| Geobacter spp./uncultured Bacteroidetes/ Cytophaga spp./ Algoriphagus spp. | 92 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Flavobacteriaceae bacterium/uncultured rumen bacterium/ Desulfovibrio spp. | 93–94 | 2 (50) | 0 (0) | 3 (50) | 4 (67) | 2 (40) | 5 (83) |
| Desulfovibrio profundus/ uncultured bacterium | 95 | 2 (50) | 0 (0) | 3 (50) | 1 (17) | 1 (20) | 0 (0) |
| Uncultured bacterium | 96 | 1 (25) | 1 (25) | 3 (50) | 4 (67) | 2 (40) | 4 (67) |
| Desulfococcus oleovorans/ Desulfomonile limimaris/ Helicobacter pylori | 97–98 | 1 (25) | 0 (0) | 4 (67) | 4 (67) | 2 (40) | 4 (67) |
| Helicobacter pylori/ uncultured rumen bacterium | 99 | 2 (50) | 1 (17) | 2 (33) | 1 (20) | 5 (83) | |
| Bacteroides spp./ uncultured rumen bacterium | 100 | 3 (75) | 3 (75) | 2 (33) | 1 (17) | 2 (40) | 0 (0) |
| Bacteroides fragilis/ uncultured rumen bacterium/ Prevotella ruminicola | 101–102 | 1 (25) | 2 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 103–104 | 1 (25) | 1 (25) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured bacterium | 105 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Desulfitobacterium hafniense | 107–108 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Thiobacillus spp. | 110–111 | 1 (25) | 2 (50) | 0 (0) | 0 (0) | 1 (20) | 0 (0) |
| Uncultured bacterium | 112 | 0 (0) | 1 (25) | 0 (0) | 1 (17) | 0 (0) | 0 (0) |
| Unidentified | 113 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 1 (17) |
| Uncultured bacterium | 116 | 2 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Unidentified | 117 | 1 (25) | 2 (50) | 4 (67) | 3 (50) | 2 (40) | 5 (83) |
| Desulfitobacterium hafniense | 118 | 1 (25) | 0 (0) | 2 (33) | 1 (17) | 1 (20) | 0 (0) |
| Unidentified | 123–124 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Unidentified | 127–129 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Unidentified | 136 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 137 | 1 (25) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Microbacterium spp. | 144–145 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) |
| Leucobacter spp./ Janibacter spp. | 147 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Unidentified | 148–149 | 0 (0) | 0 (0) | 2 (33) | 0 (0) | 1 (20) | 0 (0) |
| Pseudomonas aeruginosa | 155 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 1 (20) | 1 (17) |
| Unidentified | 156 | 1 (25) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) |
| Unidentified | 163 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 1 (20) | 1 (17) |
| Synechococcus spp. | 164 | 1 (25) | 0 (0) | 0 (0) | 3 (50) | 0 (0) | 0 (0) |
| Unidentified | 165–167 | 0 (0) | 0 (0) | 3 (50) | 0 (0) | 1 (20) | 0 (0) |
| Uncultured bacterium | 178 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 179 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 1 (17) |
| Uncultured rumen bacterium | 180 | 0 (0) | 0 (0) | 0 (0) | 2 (33) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 181–182 | 1 (25) | 0 (0) | 2 (33) | 2 (33) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 183 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Uncultured rumen bacterium | 184–185 | 1 (25) | 0 (0) | 2 (33) | 0 (0) | 1 (20) | 0 (0) |
| Listeria monocytogenes | 186 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) |
| Uncultured rumen bacterium | 187 | 1 (25) | 0 (0) | 1 (17) | 0 (0) | 1 (20) | 1 (17) |
| Uncultured rumen bacterium | 193 | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Psychrobacter spp./ uncultured bacterium/ Francisella spp. | 194–195 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Unidentified | 199 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Bacillus spp. | 200 | 0 (0) | 0 (0) | 1 (17) | 1 (17) | 0 (0) | 0 (0) |
| Clostridium rectum/ uncultured bacterium/ Mycobacterium spp. | 201 | 1 (25) | 0 (0) | 3 (50) | 0 (0) | 1 (20) | 1 (17) |
| Fervidobacterium spp./ Dehalococcoides spp./ uncultured bacterium | 202 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) |
| Uncultured rumen bacterium | 203–204 | 2 (50) | 1 (25) | 3 (50) | 1 (17) | 1 (20) | 5 (83) |
| Clostridium spp. | 231–232 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Clostridium perfringens | 234–235 | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 1 (20) | 1 (17) |
| Clostridium botulinum | 237 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 1 (17) |
| Bacillus subtilis subsp. Subtilis | 238 | 1 (25) | 0 (0) | 1 (17) | 0 (0) | 0 (0) | 0 (0) |
| Bacillus subtilis subsp. subtilis/Bacillus licheniformis/Bacillus spp. | 241–242 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Geobacillus stearothermophilus/Paenibacillus spp. | 244 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Microbispora spp./ Pseudonocardia compacta/Nonomuraea bangladeshensis/Kineosporia aurantiaca | 356 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 0 (0) |
| Microbispora spp./ Herbidospora spp. | 357 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) |
| Kribbella spp./ Actinomadura spp./ Pseudonocardia spp./ Anaplasma marginale | 358 | 0 (0) | 0 (0) | 2 (33) | 2 (33) | 0 (0) | 1 (17) |
| Arthrobacter spp. | 359 | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 0 (0) | 1 (17) |
| Uncultured bacterium | 500 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (17) |
| Probe | Primer (5′→3′) | Target | Hybridization Conditions | ||
|---|---|---|---|---|---|
| Temp (°C) | Formamide | Lysozyme | |||
| EUB338 | GCTGCCTCCCGTAGGAGT | All bacteria | 50 | ||
| NON338 | ACATCCTACGGGAGGC | Non bacteria (negative control) | 50 | ||
| BAC303 | CCAATGTGGGGGACCTT | Bacteroides spp. | 48 | ||
| EREC482 | GCTTCTTAGTCAGGTACCG | Clostridiumcoccoides cluster XIVa | 50 | ||
| LAB158 | GGTATTAGCACCTGTTTCCA | Lactobacillus- Enterococcus spp. | 50 | 90 min, 37 °C | |
| ENT-D | TGCTCTCGCGAGGTCGCTTCTCTT | Enterobacteria | 50 | ||
| VER620 | ATGTGCCGTCCGCGGGTT | Verrucobacteria | 50 | 30% | |
| BIF164 | CATCCGGCATTACCACCC | Bifidobacterium spp. | 50 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrer, M.; Aguilera, M.; Martinez, V. Effects of Rifaximin on Luminal and Wall-Adhered Gut Commensal Microbiota in Mice. Int. J. Mol. Sci. 2021, 22, 500. https://doi.org/10.3390/ijms22020500
Ferrer M, Aguilera M, Martinez V. Effects of Rifaximin on Luminal and Wall-Adhered Gut Commensal Microbiota in Mice. International Journal of Molecular Sciences. 2021; 22(2):500. https://doi.org/10.3390/ijms22020500
Chicago/Turabian StyleFerrer, Marina, Mònica Aguilera, and Vicente Martinez. 2021. "Effects of Rifaximin on Luminal and Wall-Adhered Gut Commensal Microbiota in Mice" International Journal of Molecular Sciences 22, no. 2: 500. https://doi.org/10.3390/ijms22020500






