Post-Translational Modifications of Nitrate Reductases Autoregulates Nitric Oxide Biosynthesis in Arabidopsis
Abstract
:1. Introduction
2. Results
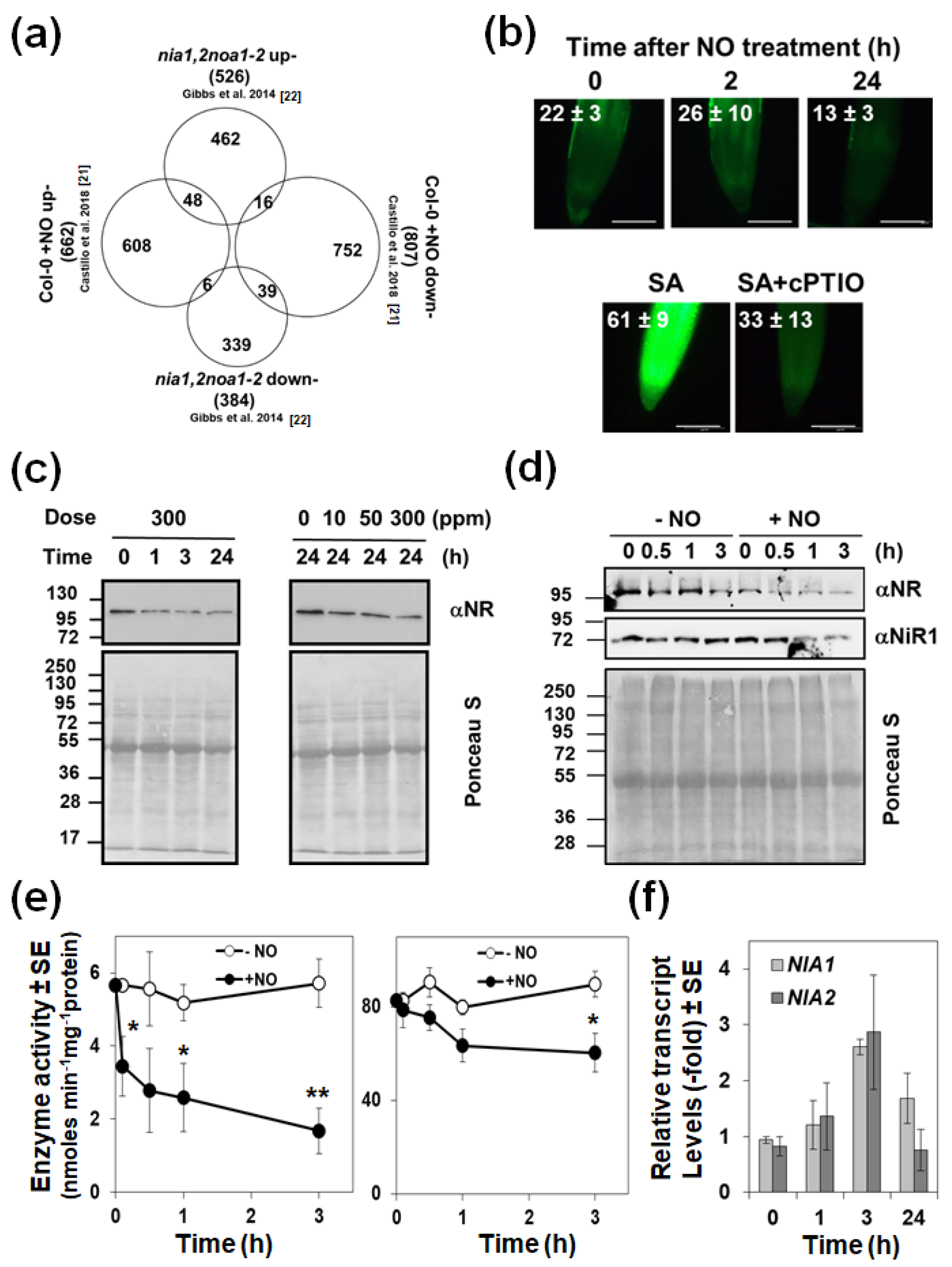
2.1. Exogenously Applied NO Represses the Endogenous Biosynthesis Acting on NRs
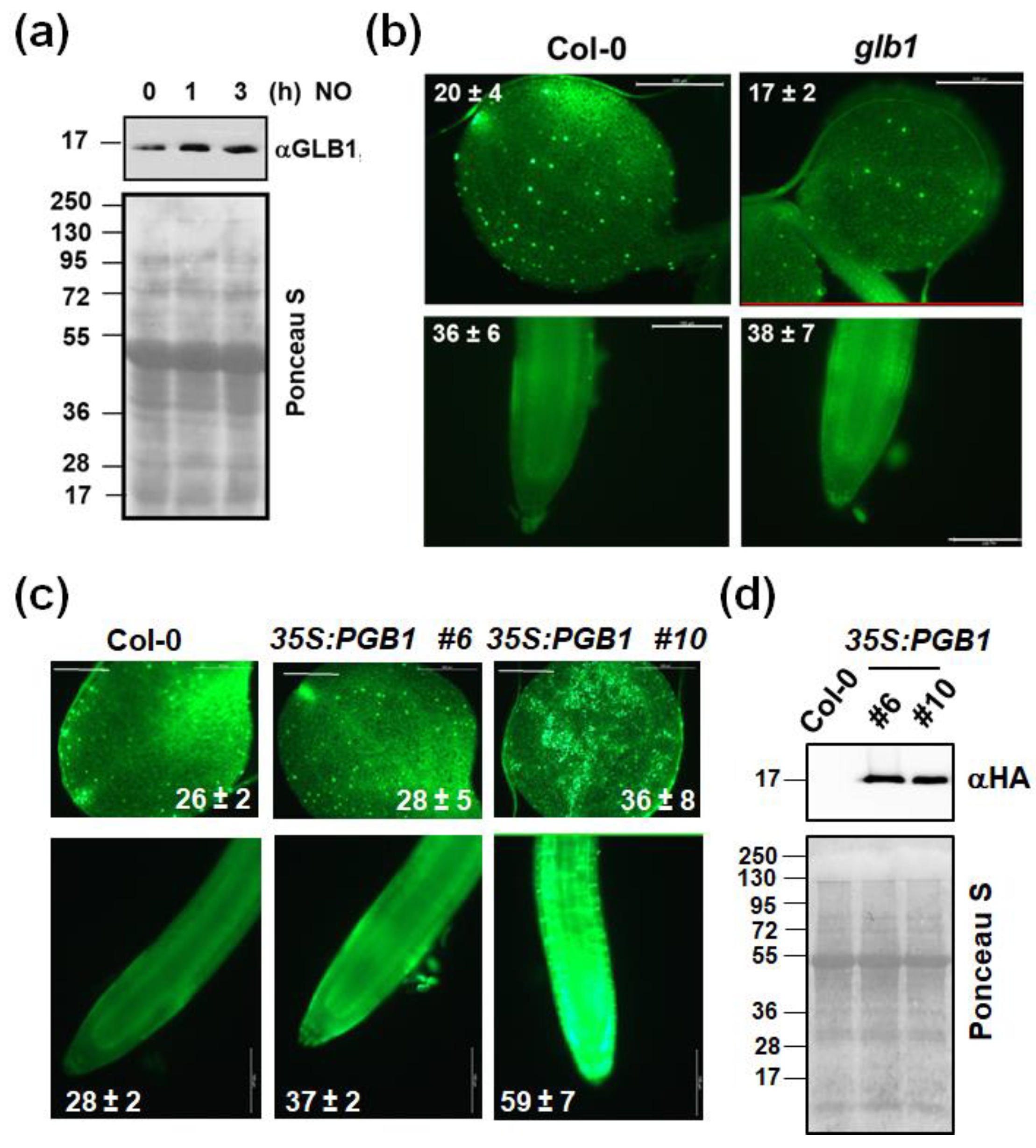
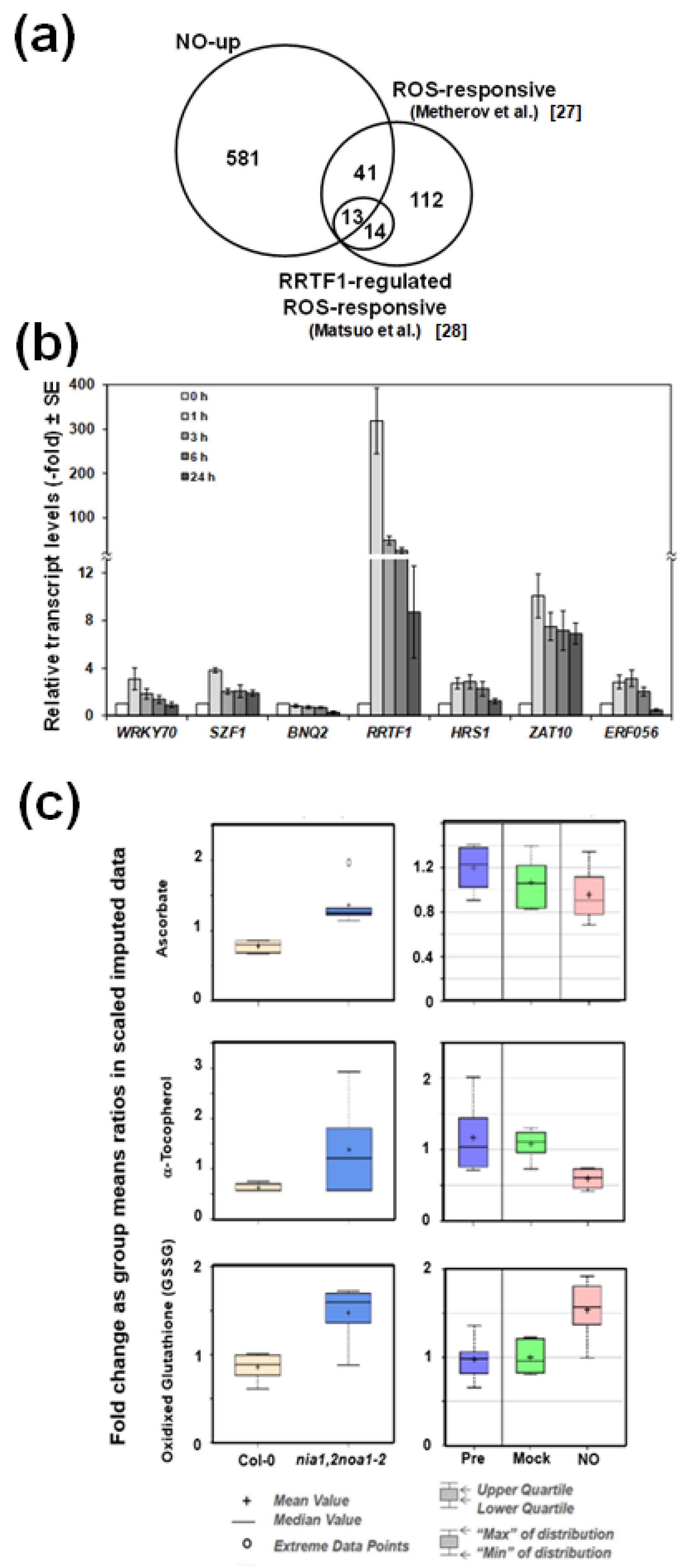
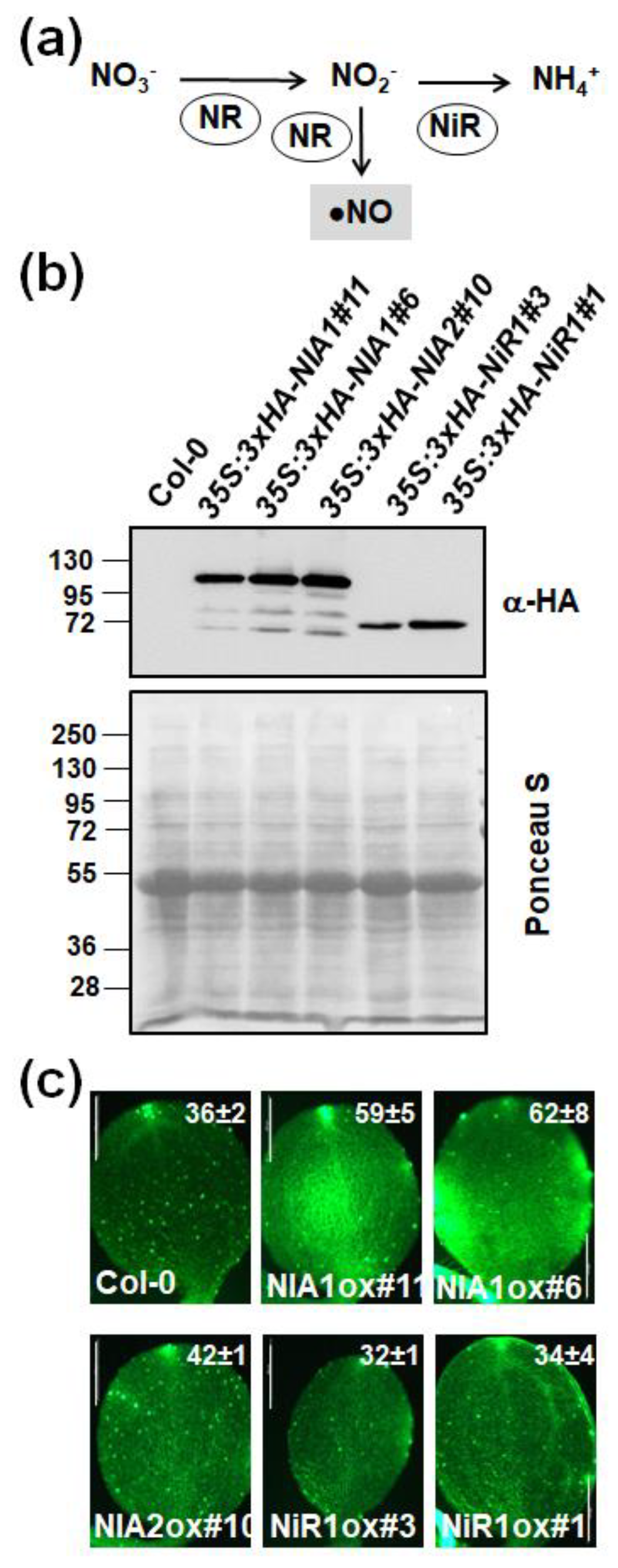
2.2. The Endogenous NO Levels Are Mainly Controlled by Post-Translational Modifications of NRs
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA iIsolation and RT-qPCR
4.3. Western Blot Analyses
4.4. Proteomic Analyses of HA-Tagged Overexpressing Plants
4.5. Antioxidant Metabolomic Analyses
4.6. Nitrate Reductase and Nitrite Reductase Activity Assays
4.7. Measurement of Endogenous NO Content
4.8. Protein Sequence Analyses and 3D Structure Modelling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hancock, J.T.; Neill, S.J. Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds. Plants 2019, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisslocker-Schaetzel, M.; André, F.; Touazi, N.; Foresi, N.; Lembrouk, M.; Dorlet, P.; Frelet-Barrand, A.; Lamattina, L.; Santolini, J. The NOS-like protein from the microalgae Ostreococcus tauri is a genuine and ultrafast NO-producing enzyme. Plant Sci. 2017, 265, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Solomonson, I.P.; Barber, M.J. Assimilatory nitrate reductase: Functional properties and regulation. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1990, 41, 225–253. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Schwarz, G. Nitrite-dependent nitric oxide synthesis by molybdenum enzymes. FEBS Lett. 2018, 592, 2126–2139. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.J.; Igamberdiev, A.U. The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 2011, 11, 537–543. [Google Scholar] [CrossRef]
- Cheng, C.L.; Dewdney, J.; Nam, H.G.; den Boer, B.G.; Goodman, H.M. A new locus (NIA 1) in Arabidopsis thaliana encoding nitrate reductase. EMBO J. 1988, 7, 3309–3314. [Google Scholar] [CrossRef]
- Wilkinson, J.Q.; Crawford, N.M. Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 1991, 3, 461–471. [Google Scholar]
- Costa-Broseta, A.; Castillo, M.C.; León, J. Nitrite reductase 1 is a target of nitric oxide-mediated post-translational modifications and controls nitrogen flux and growth in Arabidopsis. Int. J. Mol. Sci. 2020, 21, E7270. [Google Scholar] [PubMed]
- Kaiser, W.M.; Huber, S.C. Post-translational regulation of nitrate reductase: Mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 2001, 52, 1981–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lillo, C.; Meyer, C.; Lea, U.S.; Provan, F.; Oltedal, S. Mechanism and importance of post-translational regulation of nitrate reductase. J. Exp. Bot 2004, 55, 1275–1282. [Google Scholar] [PubMed]
- MacKintosh, C.; Meek, S.E. Regulation of plant NR activity by reversible phosphorylation, 14-3-3 proteins and proteolysis. Cell. Mol. Life Sci. 2001, 58, 205–214. [Google Scholar]
- Huber, J.L.; Huber, S.C.; Campbell, W.H.; Redinbaugh, M.G. Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch. Biochem. Biophys. 1992, 296, 58–65. [Google Scholar]
- Weiner, H.; Kaiser, W.M. 14-3-3 proteins control proteolysis of nitrate reductase in spinach leaves. FEBS Lett. 1999, 455, 75–78. [Google Scholar] [CrossRef] [Green Version]
- Park, B.; Song, J.T.; Seo, H.S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2011, 2, 400. [Google Scholar]
- Campbell, W.H.; Kinghorn, K.R. Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem. Sci. 1990, 15, 315–319. [Google Scholar]
- Crawford, N.M.; Smith, M.; Bellissimo, D.; Davis, R.W. Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc. Natl. Acad. Sci. USA 1988, 85, 5006–5010. [Google Scholar]
- Hyde, G.E.; Wilberding, J.A.; Meyer, A.L.; Campbell, E.R.; Campbell, W.H. Monoclonal antibody-based immunoaffinity chromatography for purifying corn and squash NADH: Nitrate reductases. Evidence for an interchain disulfide bond in nitrate reductase. Plant Mol. Biol. 1989, 13, 233–246. [Google Scholar] [CrossRef]
- Castillo, M.C.; Coego, A.; Costa-Broseta, Á.; León, J. Nitric oxide responses in Arabidopsis hypocotyls are mediated by diverse phytohormone pathways. J. Exp. Bot. 2018, 69, 5265–5278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Kalakoutskii, K.L.; Fernández, E. Chlamydomonas reinhardtii nitrate reductase complex has 105 kDa subunits in the wild-type strain and a structural mutant. Plant Sci. 1995, 105, 195–206. [Google Scholar] [CrossRef]
- Perazzolli, M.; Dominici, P.; Romero-Puertas, M.C.; Zago, E.; Zeier, J.; Sonoda, M.; Lamb, C.; Delledonne, M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 2004, 16, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Hargrove, M.; Arredondo-Peter, R. Phytoglobin: A novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on Oxygen-Binding and Sensing Proteins. F1000Research 2016, 5, 212. [Google Scholar] [PubMed] [Green Version]
- Kumar, N.; Astegno, A.; Chen, J.; Giorgetti, A.; Dominici, P. Residues in the Distal Heme Pocket of Arabidopsis Non-Symbiotic Hemoglobins: Implication for Nitrite Reductase Activity. Int. J. Mol. Sci. 2016, 17, 640. [Google Scholar] [CrossRef]
- Mehterov, N.; Balazadeh, S.; Hille, J.; Toneva, V.; Mueller-Roeber, B.; Gechev, T. Oxidative stress provokes distinct transcriptional responses in the stress-tolerant atr7 and stress-sensitive loh2 Arabidopsis thaliana mutants as revealed by multi-parallel quantitative real-time PCR analysis of ROS marker and antioxidant genes. Plant Physiol. Biochem. 2012, 59, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, M.; Johnson, J.M.; Hieno, A.; Tokizawa, M.; Nomoto, M.; Tada, Y.; Godfrey, R.; Obokata, J.; Sherameti, I.; Yamamoto, Y.Y.; et al. High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 Levels Result in Accumulation of Reactive Oxygen Species in Arabidopsis thaliana Shoots and Roots. Mol. Plant 2015, 8, 1253–1273. [Google Scholar] [CrossRef] [Green Version]
- Szarka, A.; Tomasskovics, B.; Bánhegyi, G. The ascorbate-glutathione-α-tocopherol triad in abiotic stress response. Int. J. Mol. Sci. 2012, 13, 4458–4483. [Google Scholar] [CrossRef] [Green Version]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar]
- León, J.; Costa-Broseta, Á. Present knowledge and controversies, deficiencies and misconceptions on nitric oxide synthesis, sensing and signaling in plants. Plant Cell Environ. 2020, 43, 1–15. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, B.S.; Park, S.W.; Lee, H.Y.; Song, J.T.; Seo, H.S. Nitrate Reductases Are Relocalized to the Nucleus by AtSIZ1 and Their Levels Are Negatively Regulated by COP1 and Ammonium. Int. J. Mol. Sci. 2018, 19, 1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.L.; Acedo, G.N.; Dewdney, J.; Goodman, H.M.; Conkling, M.A. Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 1991, 96, 275–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-Specific NO Synthesis by Arabidopsis thaliana Nitrate Reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.C.; Lozano-Juste, J.; González-Guzmán, M.; Rodriguez, L.; Rodriguez, P.L.; León, J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015, 8, ra89. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Davenport, S.; Le Lay, P.; Sanchez-Tamburrrino, J.P. Nitrate metabolism in tobacco leaves overexpressing Arabidopsis nitrite reductase. Plant Physiol. Biochem. 2015, 97, 96–107. [Google Scholar] [CrossRef] [Green Version]
- Blume, Y.B. A journey through a plant cytoskeleton: Hot spots in signaling and functioning. Cell Biol. Int. 2020, 44, 1262–1266. [Google Scholar] [CrossRef]
- Takahashi, M.; Sasaki, Y.; Ida, S.; Morikawa, H. Nitrite reductase gene enrichment improves assimilation of NO2− in Arabidopsis. Plant Physiol. 2001, 126, 731–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.Q.; Okamoto, M.; Crawford, N.M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 2003, 302, 100–103. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa-Broseta, Á.; Castillo, M.; León, J. Post-Translational Modifications of Nitrate Reductases Autoregulates Nitric Oxide Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 549. https://doi.org/10.3390/ijms22020549
Costa-Broseta Á, Castillo M, León J. Post-Translational Modifications of Nitrate Reductases Autoregulates Nitric Oxide Biosynthesis in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(2):549. https://doi.org/10.3390/ijms22020549
Chicago/Turabian StyleCosta-Broseta, Álvaro, MariCruz Castillo, and José León. 2021. "Post-Translational Modifications of Nitrate Reductases Autoregulates Nitric Oxide Biosynthesis in Arabidopsis" International Journal of Molecular Sciences 22, no. 2: 549. https://doi.org/10.3390/ijms22020549





