Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases
Abstract
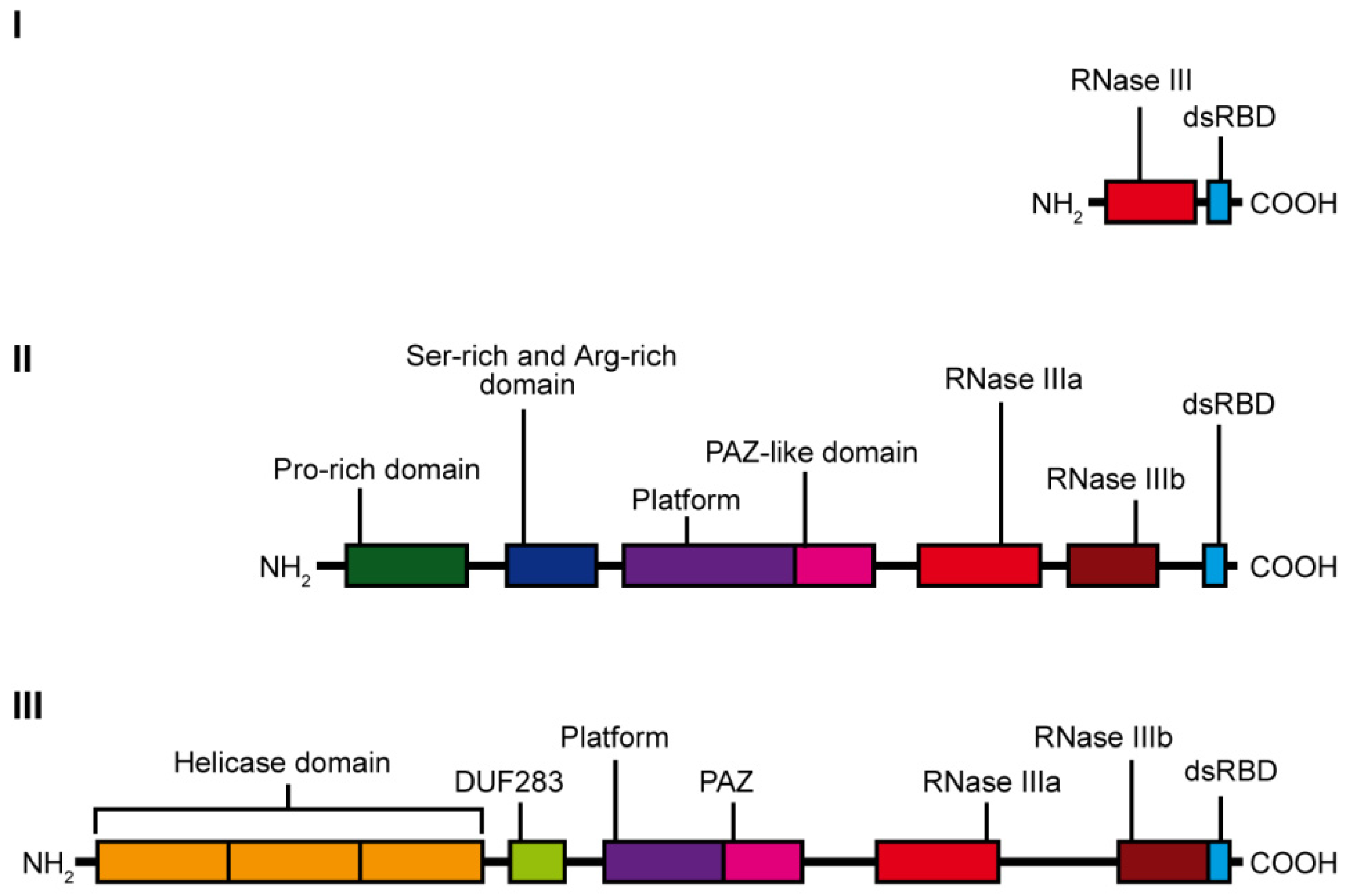
:1. Introduction
2. Evolution of Dicer-Type Proteins
3. Dicer Structure and the Importance of Its Domains
3.1. The Helicase Domain
3.2. The DUF283 Domain
3.3. Functional Core of DICER: The PAZ and Two RNase III Domains
3.4. The dsRBD Domain
4. A Variety of Dicer Functions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| ADAR | Double-stranded RNA-specific Adenosine Deaminase |
| Ago | Argonaute |
| bp | base pair |
| CAF | CARPEL FACTORY |
| cryo-EM | cryogenic Electron Microscopy |
| DCL | DICER-LIKE proteins |
| DDR | DNA Damage Response |
| ddRNA | Dicer-dependent small RNA |
| DSB | DNA Double-Strand Break |
| dsRBP | double-stranded RNA Binding Protein |
| dsRNA | double-stranded RNA |
| hDicer | human Dicer |
| HYL 1 | Hyponastic leaves 1 |
| miRNA | micro-RNA |
| NLS | Nuclear Localization Signal |
| NMR | Nuclear Magnetic Resonance |
| nt | nucleotide |
| OB fold | Oligonucleotide/Oligosaccharide-Binding fold |
| PACT | Protein Activator of the interferon-induced protein kinase |
| PPC | Platform-PAZ-Connector helix cassette |
| PPD | PAZ-Piwi Domain |
| pre-miRNA | miRNA precursor |
| pri-miRNA | miRNA primary precursor |
| RISC | RNA-induced Silencing Complex |
| SF2 | Helicase Superfamily 2 |
| siRNA | small interfering RNA |
| tasiRNA | trans-acting siRNA |
| TRBP | TAR RNA-binding protein |
| tRF | tRNA-derived Fragment |
| UTR | Untranslated Region |
References
- Hansen, S.R.; Aderounmu, A.M.; Donelick, H.M.; Bass, B.L. Dicer’s Helicase Domain: A Meeting Place for Regulatory Proteins. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L.; Hurst, S.R.; Singer, J.D. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 1994, 4, 301–314. [Google Scholar] [CrossRef]
- Provost, P.; Samuelsson, B.; Rådmark, O. Interaction of 5-lipoxygenase with cellular proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 1881–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, S.; Ichigotani, Y.; Okuda, T.; Irimura, T.; Nakatsugawa, S.; Hamaguchi, M. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta 2000, 1490, 163–169. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.E.; Running, M.P.; Meyerowitz, E.M. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 1999, 126, 5231–5243. [Google Scholar] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Axtell, M.J. MicroRNAs in Plants: Key Findings from the Early Years. Plant Cell 2019, 31, 1206–1207. [Google Scholar] [CrossRef] [Green Version]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elela, S.A.; Igel, H.; Ares, M. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 1996, 85, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Chanfreau, G.; Elela, S.A.; Ares, M.; Guthrie, C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997, 11, 2741–2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou Elela, S.; Ares, M. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998, 17, 3738–3746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanfreau, G.; Rotondo, G.; Legrain, P.; Jacquier, A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998, 17, 3726–3737. [Google Scholar] [CrossRef] [Green Version]
- Bardwell, J.C.; Régnier, P.; Chen, S.M.; Nakamura, Y.; Grunberg-Manago, M.; Court, D.L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989, 8, 3401–3407. [Google Scholar] [CrossRef]
- Zer, C.; Chanfreau, G. Regulation and surveillance of normal and 3′-extended forms of the yeast aci-reductone dioxygenase mRNA by RNase III cleavage and exonucleolytic degradation. J. Biol. Chem. 2005, 280, 28997–29003. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Chong, M.M.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Xu, H.; Miraglia, L.J.; Crooke, S.T. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000, 275, 36957–36965. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.H.; Crooke, S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011, 39, 4875–4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johanson, T.M.; Lew, A.M.; Chong, M.M. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013, 3, 130144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, A.A.; Santos, B.A.; Hnatova, S.; Bassett, A.R.; Molnar, A.; Chung, B.Y.; Baulcombe, D.C. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016, 26, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, K.; Campos, H.; Kolaczkowski, B. Evolution of animal and plant dicers: Early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 2013, 30, 627–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef] [Green Version]
- De Jong, D.; Eitel, M.; Jakob, W.; Osigus, H.J.; Hadrys, H.; Desalle, R.; Schierwater, B. Multiple dicer genes in the early-diverging metazoa. Mol. Biol. Evol. 2009, 26, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Kurzynska-Kokorniak, A.; Koralewska, N.; Pokornowska, M.; Urbanowicz, A.; Tworak, A.; Mickiewicz, A.; Figlerowicz, M. The many faces of Dicer: The complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res. 2015, 43, 4365–4380. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011, 333, 1592. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 2013, 9, e1003089. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef] [Green Version]
- Bouché, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006, 25, 3347–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Poethig, R.S.; Bowman, L.H.; Vance, V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.W.; Zilberman, D.; Xie, Z.; Johansen, L.K.; Carrington, J.C.; Jacobsen, S.E. RNA silencing genes control de novo DNA methylation. Science 2004, 303, 1336. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tworak, A.; Urbanowicz, A.; Podkowinski, J.; Kurzynska-Kokorniak, A.; Koralewska, N.; Figlerowicz, M. Six Medicago truncatula Dicer-like protein genes are expressed in plant cells and upregulated in nodules. Plant Cell Rep. 2016, 35, 1043–1052. [Google Scholar] [CrossRef]
- Liu, Q.; Feng, Y.; Zhu, Z. Dicer-like (DCL) proteins in plants. Funct. Integr. Genom. 2009, 9, 277–286. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Casas-Mollano, J.A.; Rohr, J.; Kim, E.J.; Balassa, E.; van Dijk, K.; Cerutti, H. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 2008, 179, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravind, L.; Walker, D.R.; Koonin, E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999, 27, 1223–1242. [Google Scholar] [CrossRef] [PubMed]
- Lestini, R.; Delpech, F.; Myllykallio, H. DNA replication restart and cellular dynamics of Hef helicase/nuclease protein in Haloferax volcanii. Biochimie 2015, 118, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef] [Green Version]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Park, J.E.; Heo, I.; Kim, V.N.; Patel, D.J. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Koralewska, N.; Ciechanowska, K.; Pokornowska, M.; Figlerowicz, M.; Kurzyńska-Kokorniak, A. Human ribonuclease Dicer—Structure and functions. Postepy Biochem. 2019, 65, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1549–1550. [Google Scholar] [CrossRef]
- Kidwell, M.A.; Chan, J.M.; Doudna, J.A. Evolutionarily conserved roles of the dicer helicase domain in regulating RNA interference processing. J. Biol. Chem. 2014, 289, 28352–28362. [Google Scholar] [CrossRef] [Green Version]
- Byrd, A.K.; Raney, K.D. Superfamily 2 helicases. Front. Biosci. (Landmark Ed.) 2012, 17, 2070–2088. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chang, M.; Nie, P.; Secombes, C.J. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 2009, 9, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jankowsky, E.; Fairman, M.E. RNA helicases—One fold for many functions. Curr. Opin. Struct. Biol. 2007, 17, 316–324. [Google Scholar] [CrossRef]
- Welker, N.C.; Maity, T.S.; Ye, X.; Aruscavage, P.J.; Krauchuk, A.A.; Liu, Q.; Bass, B.L. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell 2011, 41, 589–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Axtell, M.J.; Fedoroff, N.V. The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 2012, 159, 748–758. [Google Scholar] [CrossRef] [Green Version]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2017, 130, 33–44. [Google Scholar] [CrossRef]
- Colmenares, S.U.; Buker, S.M.; Buhler, M.; Dlakić, M.; Moazed, D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell 2007, 27, 449–461. [Google Scholar] [CrossRef]
- Cenik, E.S.; Fukunaga, R.; Lu, G.; Dutcher, R.; Wang, Y.; Tanaka Hall, T.M.; Zamore, P.D. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol. Cell 2011, 42, 172–184. [Google Scholar] [CrossRef] [Green Version]
- Van Rij, R.P.; Saleh, M.C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Hur, S. Helicases in Antiviral Immunity: Dual Properties as Sensors and Effectors. Trends Biochem. Sci. 2015, 40, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Kowalinski, E.; Lunardi, T.; McCarthy, A.A.; Louber, J.; Brunel, J.; Grigorov, B.; Gerlier, D.; Cusack, S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011, 147, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, E.; MacRae, I.J.; Kirsch, J.F.; Doudna, J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008, 380, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarthy, S.; Sternberg, S.H.; Kellenberger, C.A.; Doudna, J.A. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol. 2010, 404, 392–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, E.; Zhou, K.; Kidwell, M.A.; Doudna, J.A. Coordinated activities of human dicer domains in regulatory RNA processing. J. Mol. Biol. 2012, 422, 466–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, D.W.; Ma, E.; Shigematsu, H.; Cianfrocco, M.A.; Noland, C.L.; Nagayama, K.; Nogales, E.; Doudna, J.A.; Wang, H.W. Substrate-specific structural rearrangements of human Dicer. Nat. Struct. Mol. Biol. 2013, 20, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Soifer, H.S.; Sano, M.; Sakurai, K.; Chomchan, P.; Saetrom, P.; Sherman, M.A.; Collingwood, M.A.; Behlke, M.A.; Rossi, J.J. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008, 36, 6511–6522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, E.M.; Whisnant, A.W.; Kornepati, A.V.; Marshall, J.B.; Bogerd, H.P.; Cullen, B.R. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl. Acad. Sci. USA 2015, 112, E6945–E6954. [Google Scholar] [CrossRef] [Green Version]
- Welker, N.C.; Pavelec, D.M.; Nix, D.A.; Duchaine, T.F.; Kennedy, S.; Bass, B.L. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 2010, 16, 893–903. [Google Scholar] [CrossRef] [Green Version]
- Sinha, N.K.; Iwasa, J.; Shen, P.S.; Bass, B.L. Dicer uses distinct modules for recognizing dsRNA termini. Science 2018, 359, 329–334. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukunaga, R.; Han, B.W.; Hung, J.H.; Xu, J.; Weng, Z.; Zamore, P.D. Dicer Partner Proteins Tune the Length of Mature miRNAs in Flies and Mammals. Cell 2012, 151, 912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Doudna, J.A. TRBP alters human precursor microRNA processing in vitro. RNA 2012, 18, 2012–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, F.; Kalidas, S.; Smith, D.; Liu, Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 2006, 12, 1514–1520. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Park, J.K.; Jiang, F.; Liu, Y.; McKearin, D.; Liu, Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 2007, 13, 2324–2329. [Google Scholar] [CrossRef] [Green Version]
- Hartig, J.V.; Esslinger, S.; Böttcher, R.; Saito, K.; Förstemann, K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009, 28, 2932–2944. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Czech, B.; Brennecke, J.; Sachidanandam, R.; Wohlschlegel, J.A.; Perrimon, N.; Hannon, G.J. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 2009, 15, 1886–1895. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Chen, F.; Huan, X.; Machida, S.; Song, J.; Yuan, Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA 2010, 16, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Gleghorn, M.L.; Maquat, L.E. ‘Black sheep’ that don’t leave the double-stranded RNA-binding domain fold. Trends Biochem. Sci. 2014, 39, 328–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Paroo, Z.; Liu, Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J. Biol. Chem. 2007, 282, 28373–28378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dlakić, M. DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold. Bioinformatics 2006, 22, 2711–2714. [Google Scholar] [CrossRef] [PubMed]
- Kurzynska-Kokorniak, A.; Pokornowska, M.; Koralewska, N.; Hoffmann, W.; Bienkowska-Szewczyk, K.; Figlerowicz, M. Revealing a new activity of the human Dicer DUF283 domain in vitro. Sci. Rep. 2016, 6, 23989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokornowska, M.; Milewski, M.C.; Ciechanowska, K.; Szczepańska, A.; Wojnicka, M.; Radogostowicz, Z.; Figlerowicz, M.; Kurzynska-Kokorniak, A. The RNA-RNA base pairing potential of human Dicer and Ago2 proteins. Cell. Mol. Life Sci. 2020, 77, 3231–3244. [Google Scholar] [CrossRef] [Green Version]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef] [Green Version]
- Tahbaz, N.; Kolb, F.A.; Zhang, H.; Jaronczyk, K.; Filipowicz, W.; Hobman, T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004, 5, 189–194. [Google Scholar] [CrossRef] [Green Version]
- Kolb, F.A.; Zhang, H.; Jaronczyk, K.; Tahbaz, N.; Hobman, T.C.; Filipowicz, W. Human dicer: Purification, properties, and interaction with PAZ PIWI domain proteins. Methods Enzymol. 2005, 392, 316–336. [Google Scholar] [CrossRef]
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 2003, 426, 465–469. [Google Scholar] [CrossRef]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003, 10, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.B.; Ye, K.; Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004, 429, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, Y.; Kandeel, M.; Kondo, T.; Tanaka, A.; Makino, Y.; Miyamoto, N.; Shibata, A.; Ikeda, M.; Kitade, Y. Sulfonamide antibiotics inhibit RNAi by binding to human Argonaute protein 2 PAZ. Bioorg. Med. Chem. Lett. 2020, 30, 127637. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, S.K.; Fukunaga, R. Phosphate-binding pocket in Dicer-2 PAZ domain for high-fidelity siRNA production. Proc. Natl. Acad. Sci. USA 2016, 113, 14031–14036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Shimizu, N. Evolutionary conservation of a unique amino acid sequence in human DICER protein essential for binding to Argonaute family proteins. Gene 2007, 396, 312–320. [Google Scholar] [CrossRef]
- Wang, H.W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Tschudi, C.; Ullu, E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA 2006, 12, 2063–2072. [Google Scholar] [CrossRef] [Green Version]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, D.E.; Nakanishi, K.; Patel, D.J.; Bartel, D.P. The inside-out mechanism of Dicers from budding yeasts. Cell 2011, 146, 262–276. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Li, X.; Zheng, L.; Ma, J.; Gan, J. Structural and functional studies of a noncanonical Dicer from Entamoeba histolytica. Sci. Rep. 2017, 7, 44832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacRae, I.J.; Doudna, J.A. Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007, 17, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Mickiewicz, A.; Sarzyńska, J.; Miłostan, M.; Kurzyńska-Kokorniak, A.; Rybarczyk, A.; Łukasiak, P.; Kuliński, T.; Figlerowicz, M.; Błażewicz, J. Modeling of the catalytic core of Arabidopsis thaliana Dicer-like 4 protein and its complex with double-stranded RNA. Comput. Biol. Chem. 2017, 66, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Wojnicka, M.; Szczepanska, A.; Kurzynska-Kokorniak, A. Unknown Areas of Activity of Human Ribonuclease Dicer: A Putative Deoxyribonuclease Activity. Molecules 2020, 25, 1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dincbas-Renqvist, V.; Pépin, G.; Rakonjac, M.; Plante, I.; Ouellet, D.L.; Hermansson, A.; Goulet, I.; Doucet, J.; Samuelsson, B.; Rådmark, O.; et al. Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim. Biophys. Acta 2009, 1789, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Song, H.; Tropea, J.E.; Needle, D.; Waugh, D.S.; Gu, S.; Ji, X. The molecular mechanism of dsRNA processing by a bacterial Dicer. Nucleic Acids Res. 2019, 47, 4707–4720. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Mateos, J.L.; Bologna, N.G.; Chorostecki, U.; Palatnik, J.F. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010, 20, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Zhou, Y.; Castillo-González, C.; Lu, A.; Ge, C.; Zhao, Y.T.; Duan, L.; Li, Z.; Axtell, M.J.; Wang, X.J.; et al. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 2013, 20, 1106–1115. [Google Scholar] [CrossRef]
- Bologna, N.G.; Schapire, A.L.; Zhai, J.; Chorostecki, U.; Boisbouvier, J.; Meyers, B.C.; Palatnik, J.F. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013, 23, 1675–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chorostecki, U.; Moro, B.; Rojas, A.M.L.; Debernardi, J.M.; Schapire, A.L.; Notredame, C.; Palatnik, J.F. Evolutionary Footprints Reveal Insights into Plant MicroRNA Biogenesis. Plant Cell 2017, 29, 1248–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addo-Quaye, C.; Snyder, J.A.; Park, Y.B.; Li, Y.F.; Sunkar, R.; Axtell, M.J. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 2009, 15, 2112–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 2014, 5, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.; Rauhut, R. Ribonuclease III: New sense from nuisance. Int. J. Biochem. Cell Biol. 2002, 34, 116–129. [Google Scholar] [CrossRef]
- Blaszczyk, J.; Gan, J.; Tropea, J.E.; Court, D.L.; Waugh, D.S.; Ji, X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure 2004, 12, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Gan, J.; Tropea, J.E.; Austin, B.P.; Court, D.L.; Waugh, D.S.; Ji, X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 2006, 124, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Tyczewska, A.; Kurzyńska-Kokorniak, A.; Koralewska, N.; Szopa, A.; Kietrys, A.M.; Wrzesiński, J.; Twardowski, T.; Figlerowicz, M. Selection of RNA oligonucleotides that can modulate human dicer activity in vitro. Nucleic Acid Ther. 2011, 21, 333–346. [Google Scholar] [CrossRef]
- Kurzynska-Kokorniak, A.; Koralewska, N.; Tyczewska, A.; Twardowski, T.; Figlerowicz, M. A new short oligonucleotide-based strategy for the precursor-specific regulation of microRNA processing by dicer. PLoS ONE 2013, 8, e77703. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Pertzev, A.; Nicholson, A.W. Catalytic mechanism of Escherichia coli ribonuclease III: Kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis. Nucleic Acids Res. 2005, 33, 807–815. [Google Scholar] [CrossRef] [Green Version]
- Kerner, P.; Degnan, S.M.; Marchand, L.; Degnan, B.M.; Vervoort, M. Evolution of RNA-binding proteins in animals: Insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol. Biol. Evol. 2011, 28, 2289–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masliah, G.; Barraud, P.; Allain, F.H. RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Cell. Mol. Life Sci. 2013, 70, 1875–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.; Grünert, S.; Adams, J.; Micklem, D.R.; Proctor, M.R.; Freund, S.; Bycroft, M.; St Johnston, D.; Varani, G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000, 19, 997–1009. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Henras, A.; Chanfreau, G.; Feigon, J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. USA 2004, 101, 8307–8312. [Google Scholar] [CrossRef] [Green Version]
- Sinkkonen, L.; Hugenschmidt, T.; Filipowicz, W.; Svoboda, P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE 2010, 5, e12175. [Google Scholar] [CrossRef] [Green Version]
- Yadav, R.P.; Mäkelä, J.A.; Hyssälä, H.; Cisneros-Montalvo, S.; Kotaja, N. DICER regulates the expression of major satellite repeat transcripts and meiotic chromosome segregation during spermatogenesis. Nucleic Acids Res. 2020, 48, 7135–7153. [Google Scholar] [CrossRef]
- Doyle, M.; Badertscher, L.; Jaskiewicz, L.; Güttinger, S.; Jurado, S.; Hugenschmidt, T.; Kutay, U.; Filipowicz, W. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA 2013, 19, 1238–1252. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Barraud, P. Functions of double-stranded RNA-binding domains in nucleocytoplasmic transport. RNA Biol. 2014, 11, 1226–1232. [Google Scholar] [CrossRef] [Green Version]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef] [Green Version]
- Woolcock, K.J.; Stunnenberg, R.; Gaidatzis, D.; Hotz, H.R.; Emmerth, S.; Barraud, P.; Bühler, M. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 2012, 26, 683–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, C.S.; Vicentini, R.; Duarte, G.T.; Pinoti, V.F.; Vincentz, M.; Nogueira, F.T. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol. Biol. 2017, 93, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Grewal, S.I. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr. Opin. Genet. Dev. 2012, 22, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [Green Version]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Chen, X.; Li, W.F.; Wu, X.; Zhang, H.C.; Chen, L.; Zhang, P.Y.; Liu, L.Y.; Ma, D.; Chen, T.; Zhou, L.; et al. Dicer regulates non-homologous end joining and is associated with chemosensitivity in colon cancer patients. Carcinogenesis 2017, 38, 873–882. [Google Scholar] [CrossRef]
- Yao, Y.; Bilichak, A.; Golubov, A.; Kovalchuk, I. Arabidopsis thaliana siRNA biogenesis mutants have the lower frequency of homologous recombination. Plant Signal. Behav. 2016, 11, e1151599. [Google Scholar] [CrossRef] [Green Version]
- Burger, K.; Schlackow, M.; Potts, M.; Hester, S.; Mohammed, S.; Gullerova, M. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J. Cell Biol. 2017, 216, 2373–2389. [Google Scholar] [CrossRef]
- Chitale, S.; Richly, H. DICER- and MMSET-catalyzed H4K20me2 recruits the nucleotide excision repair factor XPA to DNA damage sites. J. Cell Biol. 2018, 217, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Chitale, S.; Richly, H. DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 2017, 45, 5901–5912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, A.; Shi, Y.; Kage-Nakadai, E.; Mitani, S.; Xue, D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 2010, 328, 327–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.W.; Kim, Y.S.; Park, J.Y.; Chu, G.E.; Yang, Y.C.; Choi, B.Y.; Cho, W.G. Hypoxia-induced apoptosis of astrocytes is mediated by reduction of Dicer and activation of caspase-1. Cell Biol. Int. 2020, 44, 1394–1404. [Google Scholar] [CrossRef]
- Koralewska, N.; Hoffmann, W.; Pokornowska, M.; Milewski, M.; Lipinska, A.; Bienkowska-Szewczyk, K.; Figlerowicz, M.; Kurzynska-Kokorniak, A. How short RNAs impact the human ribonuclease Dicer activity: Putative regulatory feedback-loops and other RNA-mediated mechanisms controlling microRNA processing. Acta Biochim. Pol. 2016, 63, 773–783. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciechanowska, K.; Pokornowska, M.; Kurzyńska-Kokorniak, A. Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. Int. J. Mol. Sci. 2021, 22, 616. https://doi.org/10.3390/ijms22020616
Ciechanowska K, Pokornowska M, Kurzyńska-Kokorniak A. Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. International Journal of Molecular Sciences. 2021; 22(2):616. https://doi.org/10.3390/ijms22020616
Chicago/Turabian StyleCiechanowska, Kinga, Maria Pokornowska, and Anna Kurzyńska-Kokorniak. 2021. "Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases" International Journal of Molecular Sciences 22, no. 2: 616. https://doi.org/10.3390/ijms22020616
APA StyleCiechanowska, K., Pokornowska, M., & Kurzyńska-Kokorniak, A. (2021). Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. International Journal of Molecular Sciences, 22(2), 616. https://doi.org/10.3390/ijms22020616





