CXCR4 Regulates Temporal Differentiation via PRC1 Complex in Organogenesis of Epithelial Glands
Abstract
:1. Introduction
2. Results
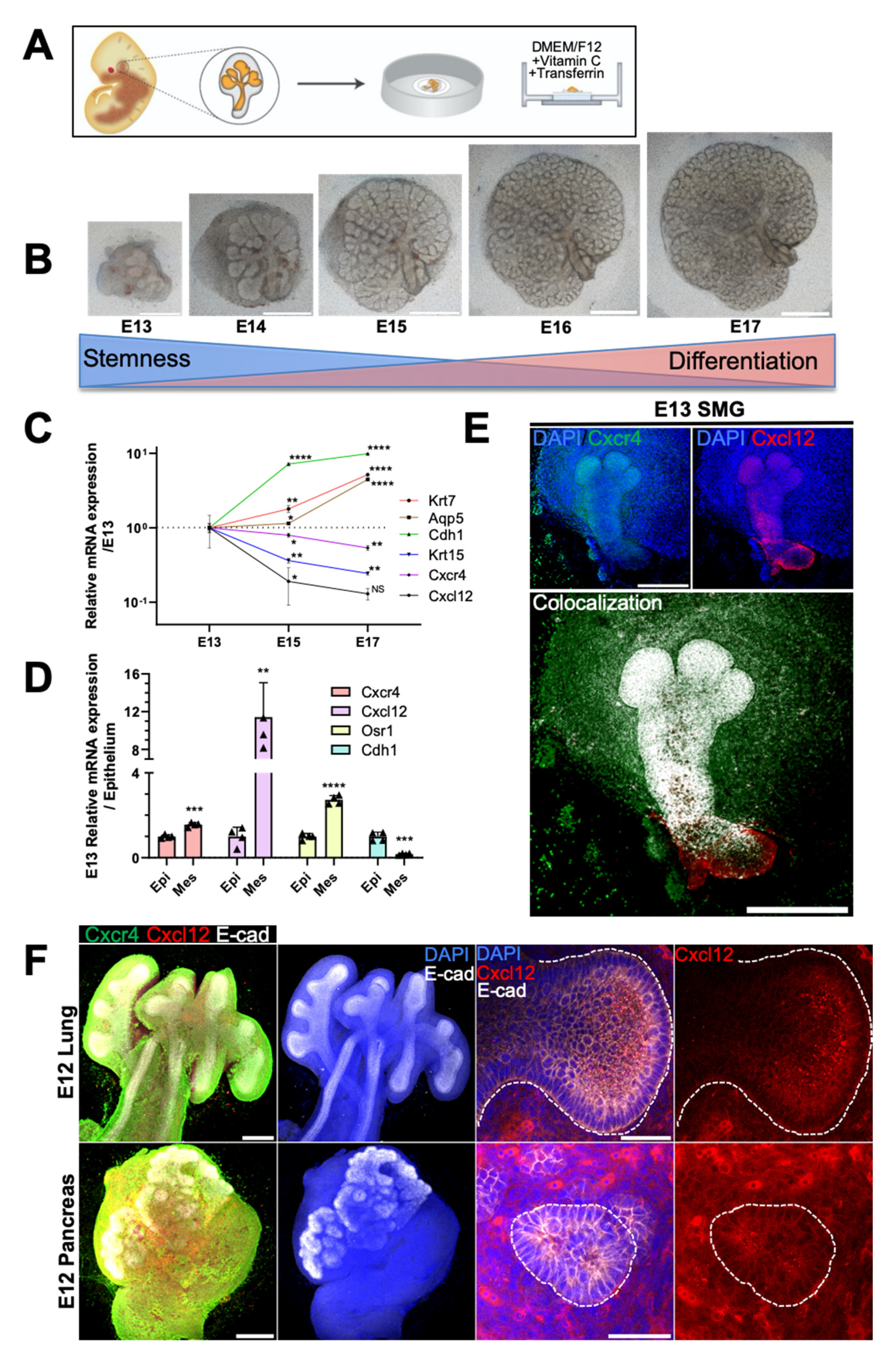
2.1. Spatiotemporal Dynamics of CXCR4 and CXCL12 Expression during Branching Morphogenesis of Epithelial Organs
2.2. AMD3100 Perturbs Glandular Organogenesis without Apoptosis
2.3. AMD3100 Opposingly Regulates Proliferation and Differentiation in Developing eSMGs
2.4. AMD3100 Alters Expressions of Developmental and PRC1-Comprising Genes in eSMGs
2.5. PRC1 Perturbation Induces Precocious Differentiation in Acinar and Ductal Cells of eSMGs
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Ex Vivo Culture of eSMGs
4.3. Epithelial Rudiment Culture
4.4. Real-Time Live Imaging
4.5. Immunofluorescence Staining and Imaging
4.6. EdU Staining
4.7. RNA Isolation and RNA-Seq Data Analysis
4.8. qRT-PCR
4.9. DNA Extraction and Methyl-Seq
4.10. Statistical Analysis
4.11. Data Deposition
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jo, D.-Y.; Rafii, S.; Hamada, T.; Moore, M.A.S. Chemotaxis of primitive hematopoietic cells in response to stromal cell–derived factor-1. J. Clin. Investig. 2000, 105, 101–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleul, C.C.; Wu, L.; Hoxie, J.A.; Springer, T.A.; Mackay, C.R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 1997, 94, 1925–1930. [Google Scholar] [CrossRef] [Green Version]
- Kurita, K.; Ishikawa, K.; Takeda, K.; Fujimoto, M.; Ono, H.; Kumagai, J.; Inoue, H.; Yokoh, H.; Yokote, K. CXCL12-CXCR4 pathway activates brown adipocytes and induces insulin resistance in CXCR4-deficient mice under high-fat diet. Sci. Rep. 2019, 9, 6165. [Google Scholar] [CrossRef]
- Pelekanos, R.A.; Ting, M.J.; Sardesai, V.S.; Ryan, J.M.; Lim, Y.-C.; Chan, J.K.Y.; Fisk, N.M. Intracellular trafficking and endocytosis of CXCR4 in fetal mesenchymal stem/stromal cells. BMC Cell Biol. 2014, 15, 15. [Google Scholar] [CrossRef] [Green Version]
- Ratajczak, M.; Zuba-Surma, E.; Kucia, M.; Reca, R.; Wojakowski, W.; Ratajczak, J. The pleiotropic effects of the SDF-1–CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia 2006, 20, 1915–1924. [Google Scholar] [CrossRef] [Green Version]
- Katsumoto, K.; Kume, S. The role of CXCL12-CXCR4 signaling pathway in pancreatic development. Theranostics 2013, 3, 11–17. [Google Scholar] [CrossRef]
- Song, Z.-Y.; Wang, F.; Cui, S.-X.; Qu, X.-J. Knockdown of CXCR4 inhibits CXCL12-induced angiogenesis in HUVECs through downregulation of the MAPK/ERK and PI3K/AKT and the Wnt/β-catenin pathways. Cancer Investig. 2018, 36, 10–18. [Google Scholar] [CrossRef]
- Takabatake, Y.; Sugiyama, T.; Kohara, H.; Matsusaka, T.; Kurihara, H.; Koni, P.A.; Nagasawa, Y.; Hamano, T.; Matsui, I.; Kawada, N.; et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J. Am. Soc. Nephrol. 2009, 20, 1714–1723. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, K.; Hirota, S.; Iizasa, H.; Yoshida, H.; Kawabata, K.; Kataoka, Y.; Kitamura, Y.; Matsushima, K.; Yoshida, N.; Nishikawa, S.-i.; et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 1998, 393, 591–594. [Google Scholar] [CrossRef] [PubMed]
- LaRocca, T.J.; Altman, P.; Jarrah, A.A.; Gordon, R.; Wang, E.; Hadri, L.; Burke, M.W.; Haddad, G.E.; Hajjar, R.J.; Tarzami, S.T. CXCR4 cardiac specific knockout mice develop a progressive cardiomyopathy. Int. J. Mol. Sci. 2019, 20, 2267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerman, N.P.; Vongsa, R.A.; Faherty, S.L.; Salzman, N.H.; Dwinell, M.B. Targeted intestinal epithelial deletion of the chemokine receptor CXCR4 reveals important roles for extracellular-regulated kinase-1/2 in restitution. Lab. Investig. 2011, 91, 1040–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribatti, D.; Santoiemma, M. Epithelial-mesenchymal interactions: A fundamental Developmental Biology mechanism. Int. J. Dev. Biol. 2014, 58, 303–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Hu, Y.; Fan, X.; Wu, X.; Mao, Y.; Hu, B.; Guo, H.; Wen, L.; Tang, F. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 2018, 19, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landsman, L.; Nijagal, A.; Whitchurch, T.J.; Vanderlaan, R.L.; Zimmer, W.E.; MacKenzie, T.C.; Hebrok, M. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biol. 2011, 9, e1001143. [Google Scholar] [CrossRef] [Green Version]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Fathi, A.; Eisa-Beygi, S.; Baharvand, H. Signaling molecules governing pluripotency and early lineage commitments in human pluripotent stem cells. Cell J. 2017, 19, 194–203. [Google Scholar] [CrossRef]
- Chatzeli, L.; Gaete, M.; Tucker, A.S. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 2017, 144, 2294–2305. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540. [Google Scholar] [CrossRef]
- Hohwieler, M.; Illing, A.; Hermann, P.C.; Mayer, T.; Stockmann, M.; Perkhofer, L.; Eiseler, T.; Antony, J.S.; Müller, M.; Renz, S.; et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 2017, 66, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Schutgens, F.; Verhaar, M.C.; Rookmaaker, M.B. Pluripotent stem cell-derived kidney organoids: An in vivo-like in vitro technology. Eur. J. Pharmacol. 2016, 790, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, J.; Ogawa, M.; Hojo, H.; Kawashima, Y.; Mabuchi, Y.; Hata, K.; Nakamura, S.; Yasuhara, R.; Takamatsu, K.; Irié, T.; et al. Generation of orthotopically functional salivary gland from embryonic stem cells. Nat. Commun. 2018, 9, 4216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajasekhar, V.K.; Begemann, M. Concise review: Roles of polycomb group proteins in development and disease: A stem cell perspective. Stem Cells 2007, 25, 2498–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chittock, E.C.; Latwiel, S.; Miller, T.C.R.; Müller, C.W. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017, 45, 193–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knosp, W.M.; Knox, S.M.; Hoffman, M.P. Salivary gland organogenesis. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, E.; May, A.J.; Nathan, S.; Cruz-Pacheco, N.; Lizama, C.O.; Maliskova, L.; Zovein, A.C.; Shen, Y.; Muench, M.O.; Knox, S.M. SOX2 regulates acinar cell development in the salivary gland. eLife 2017, 6, e26620. [Google Scholar] [CrossRef] [PubMed]
- Knox, S.M.; Lombaert, I.M.A.; Reed, X.; Vitale-Cross, L.; Gutkind, J.S.; Hoffman, M.P. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 2010, 329, 1645–1647. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-w.; Kim, J.; Do, M.; Namkoong, E.; Lee, H.; Ryu, J.H.; Park, K. Developmental role of hyaluronic acid and its application in salivary gland tissue engineering. Acta Biomater. 2020, 115, 275–287. [Google Scholar] [CrossRef]
- De Clercq, E. The AMD3100 story: The path to the discovery of a stem cell mobilizer (Mozobil). Biochem. Pharmacol. 2009, 77, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Michowski, W.; Kolodziejczyk, A.; Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 2019, 21, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Nedvetsky, P.I.; Emmerson, E.; Finley, J.K.; Ettinger, A.; Cruz-Pacheco, N.; Prochazka, J.; Haddox, C.L.; Northrup, E.; Hodges, C.; Mostov, K.E.; et al. Parasympathetic innervation regulates tubulogenesis in the developing salivary gland. Dev. Cell 2014, 30, 449–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogl, C.; Mohammed, F.; Al-Jassar, C.; Jeeves, M.; Knowles, T.J.; Rodriguez-Zamora, P.; White, S.A.; Odintsova, E.; Overduin, M.; Chidgey, M. Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin. Nat. Commun. 2016, 7, 10827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, K.J.; Crisp, M.L.; Liu, Q.; Kim, D.; Kozlov, S.; Stewart, C.L.; Burke, B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA 2009, 106, 2194–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, J.A.; Kingston, R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endoh, M.; Endo, T.A.; Endoh, T.; Isono, K.-i.; Sharif, J.; Ohara, O.; Toyoda, T.; Ito, T.; Eskeland, R.; Bickmore, W.A.; et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012, 8, e1002774. [Google Scholar] [CrossRef] [Green Version]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell 2020, 77, 840–856. [Google Scholar] [CrossRef] [Green Version]
- Gertych, A.; Oh, J.H.; Wawrowsky, K.A.; Weisenberger, D.J.; Tajbakhsh, J. 3-D DNA methylation phenotypes correlate with cytotoxicity levels in prostate and liver cancer cell models. BMC Pharmacol. Toxicol. 2013, 14, 11. [Google Scholar] [CrossRef] [Green Version]
- Milosevich, N.; Warmerdam, Z.; Hof, F. Structural aspects of small-molecule inhibition of methyllysine reader proteins. Future Med. Chem. 2016, 8, 1681–1702. [Google Scholar] [CrossRef]
- Levoye, A.; Balabanian, K.; Baleux, F.; Bachelerie, F.; Lagane, B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 2009, 113, 6085–6093. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, E.L.; Lee, W.; Lu, J.; Lowy, A.M.; Kim, J. Chemokine CXCL12 activates dual CXCR4 and CXCR7-mediated signaling pathways in pancreatic cancer cells. J. Transl. Med. 2012, 10, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-W.; Kim, D.-M.; Jang, J.-W.; Park, T.-G.; Song, S.-H.; Lee, Y.-S.; Chi, X.-Z.; Park, I.Y.; Hyun, J.-W.; Ito, Y.; et al. RUNX3 regulates cell cycle-dependent chromatin dynamics by functioning as a pioneer factor of the restriction-point. Nat. Commun. 2019, 10, 1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosogane, M.; Funayama, R.; Nishida, Y.; Nagashima, T.; Nakayama, K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 2013, 9, e1003698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.R.; Larsen, M. The contribution of specific cell subpopulations to submandibular salivary gland branching morphogenesis. Curr. Opin. Genet. Dev. 2015, 32, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loubiere, V.; Martinez, A.-M.; Cavalli, G. Cell fate and developmental regulation dynamics by polycomb proteins and 3D genome architecture. BioEssays 2019, 41, 1800222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, R.-g.; Zhang, Y.; Sun, T.-t.; Cheng, B. Epigenetic regulation by polycomb group complexes: Focus on roles of CBX proteins. J. Zhejiang Univ. Sci. B 2014, 15, 412–428. [Google Scholar] [CrossRef] [Green Version]
- Connelly, K.E.; Weaver, T.M.; Alpsoy, A.; Gu, B.X.; Musselman, C.A.; Dykhuizen, E.C. Engagement of DNA and H3K27me3 by the CBX8 chromodomain drives chromatin association. Nucleic Acids Res. 2019, 47, 2289–2305. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.; Choi, S.-P.; Isono, K.; Lee, J.Y.; Park, S.-W.; Choi, C.-Y.; Han, J.; Kim, S.-H.; Lee, H.-H.; Park, K.; et al. Phc2 controls hematopoietic stem and progenitor cell mobilization from bone marrow by repressing Vcam1 expression. Nat. Commun. 2019, 10, 3496. [Google Scholar] [CrossRef]
- Endoh, M.; Endo, T.A.; Endoh, T.; Fujimura, Y.-i.; Ohara, O.; Toyoda, T.; Otte, A.P.; Okano, M.; Brockdorff, N.; Vidal, M.; et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 2008, 135, 1513–1524. [Google Scholar] [CrossRef] [Green Version]
- Dahlet, T.; Lleida, A.A.; Al Adhami, H.; Dumas, M.; Bender, A.; Ngondo, R.P.; Tanguy, M.; Vallet, J.; Auclair, G.; Bardet, A.F.; et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat. Commun. 2020, 11, 3135. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, T.; Lombaert, I.M.A.; Hauser, B.R.; Patel, V.N.; Hoffman, M.P. Exosomal microRNA transport from salivary mesenchyme regulates epithelial progenitor expansion during organogenesis. Dev. Cell 2017, 40, 95–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.; Lee, S.-W.; Namkoong, E.; An, W.; Lee, J.-H.; Brown, P.D.; Park, K. Epigenetic modification as a regulatory mechanism for spatiotemporal dynamics of ANO1 expression in salivary glands. Int. J. Mol. Sci. 2019, 20, 6298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.M.; Choi, S.; Lee, S.W.; Park, K. Voltage-dependent Ca2+ channels promote branching morphogenesis of salivary glands by patterning differential growth. Sci. Rep. 2018, 8, 7566. [Google Scholar] [CrossRef]
- Keil, K.P.; Abler, L.L.; Mehta, V.; Altmann, H.M.; Laporta, J.; Plisch, E.H.; Suresh, M.; Hernandez, L.L.; Vezina, C.M. DNA methylation of E-cadherin is a priming mechanism for prostate development. Dev. Biol. 2014, 387, 142–153. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Larsen, M.; Yamada, K.M. Fibronectin requirement in branching morphogenesis. Nature 2003, 423, 876–881. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, M.E.; Mezzapelle, R. The chemokine receptor CXCR4 in cell proliferation and tissue regeneration. Front. Immunol. 2020, 11, 2109. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lee, S.-W.; Park, K. CXCR4 Regulates Temporal Differentiation via PRC1 Complex in Organogenesis of Epithelial Glands. Int. J. Mol. Sci. 2021, 22, 619. https://doi.org/10.3390/ijms22020619
Kim J, Lee S-W, Park K. CXCR4 Regulates Temporal Differentiation via PRC1 Complex in Organogenesis of Epithelial Glands. International Journal of Molecular Sciences. 2021; 22(2):619. https://doi.org/10.3390/ijms22020619
Chicago/Turabian StyleKim, Junchul, Sang-Woo Lee, and Kyungpyo Park. 2021. "CXCR4 Regulates Temporal Differentiation via PRC1 Complex in Organogenesis of Epithelial Glands" International Journal of Molecular Sciences 22, no. 2: 619. https://doi.org/10.3390/ijms22020619




