Intracellular Ionic Strength Sensing Using NanoLuc
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Luciferase Assays
2.2. In Silico Docking and Simulation
2.3. Cell Lysate and Live-Cell Assays
2.4. Cell Culture and Transfection
2.5. Data Analysis and Figure Preparation
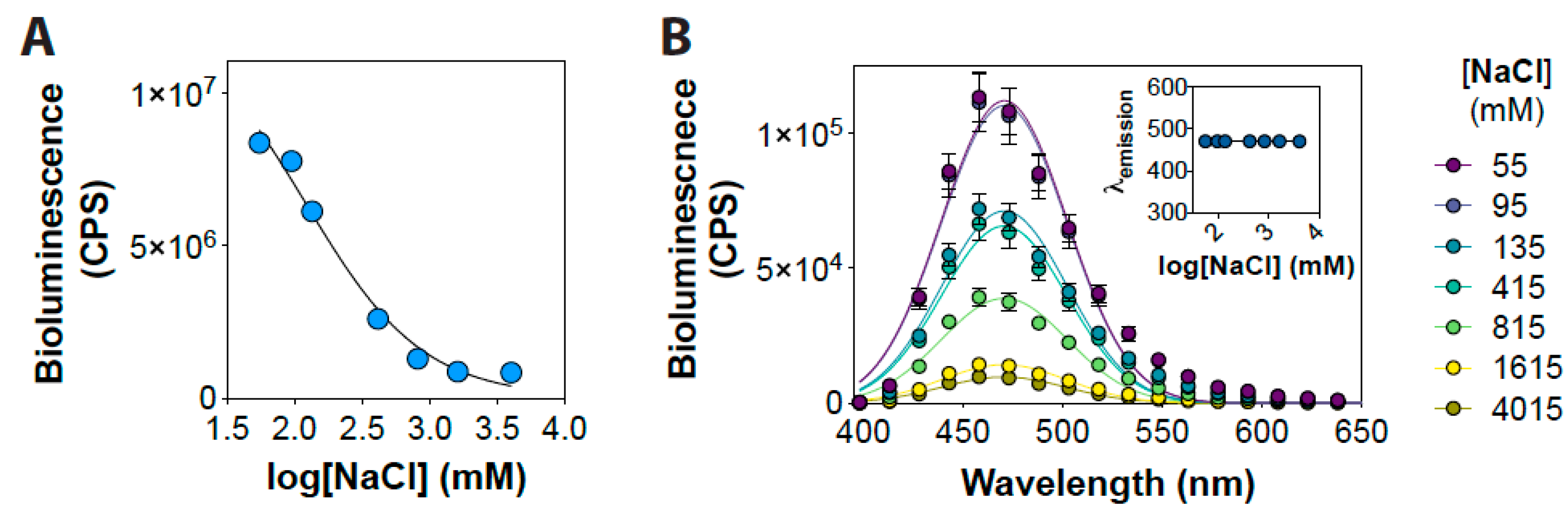
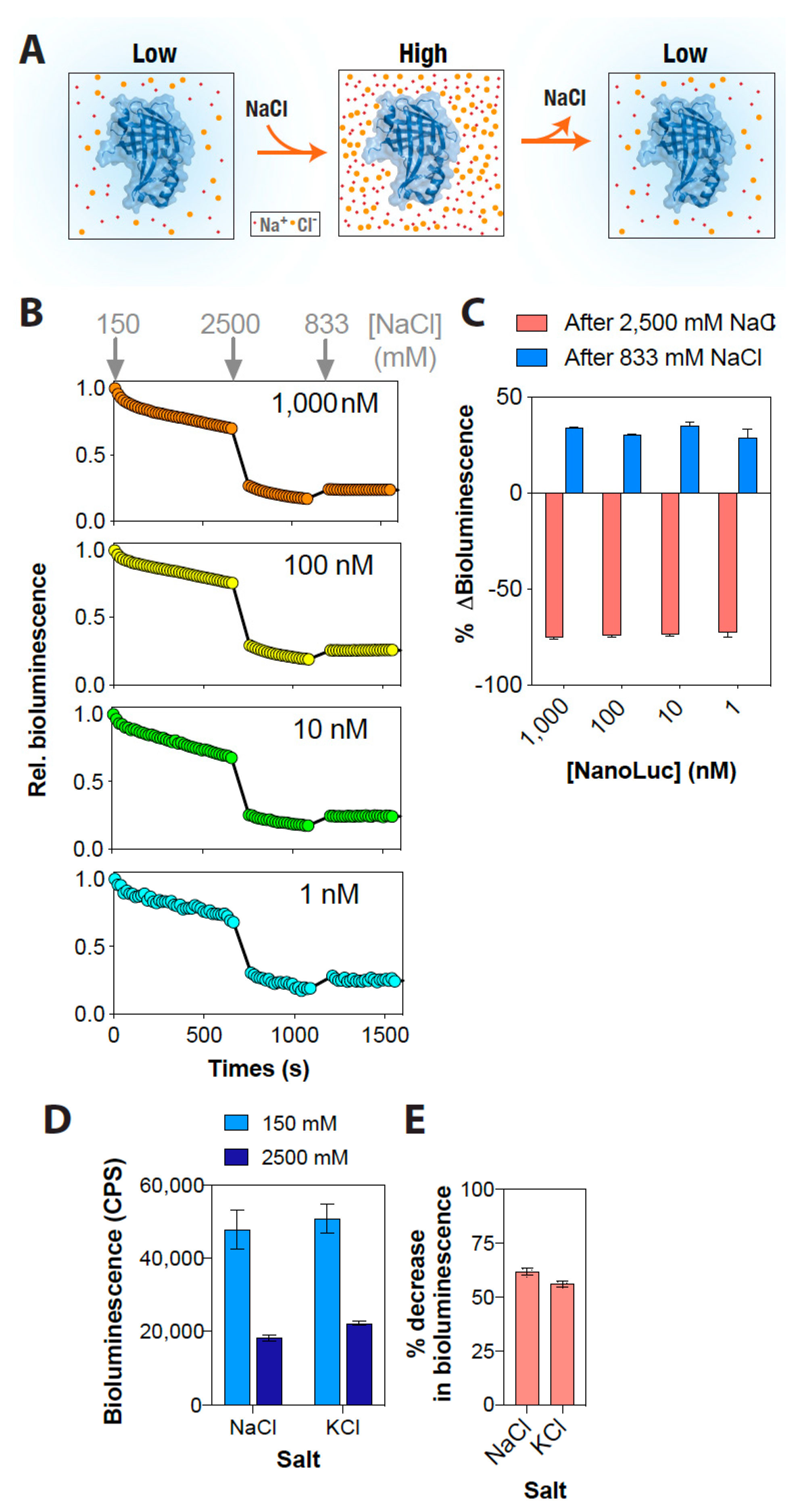
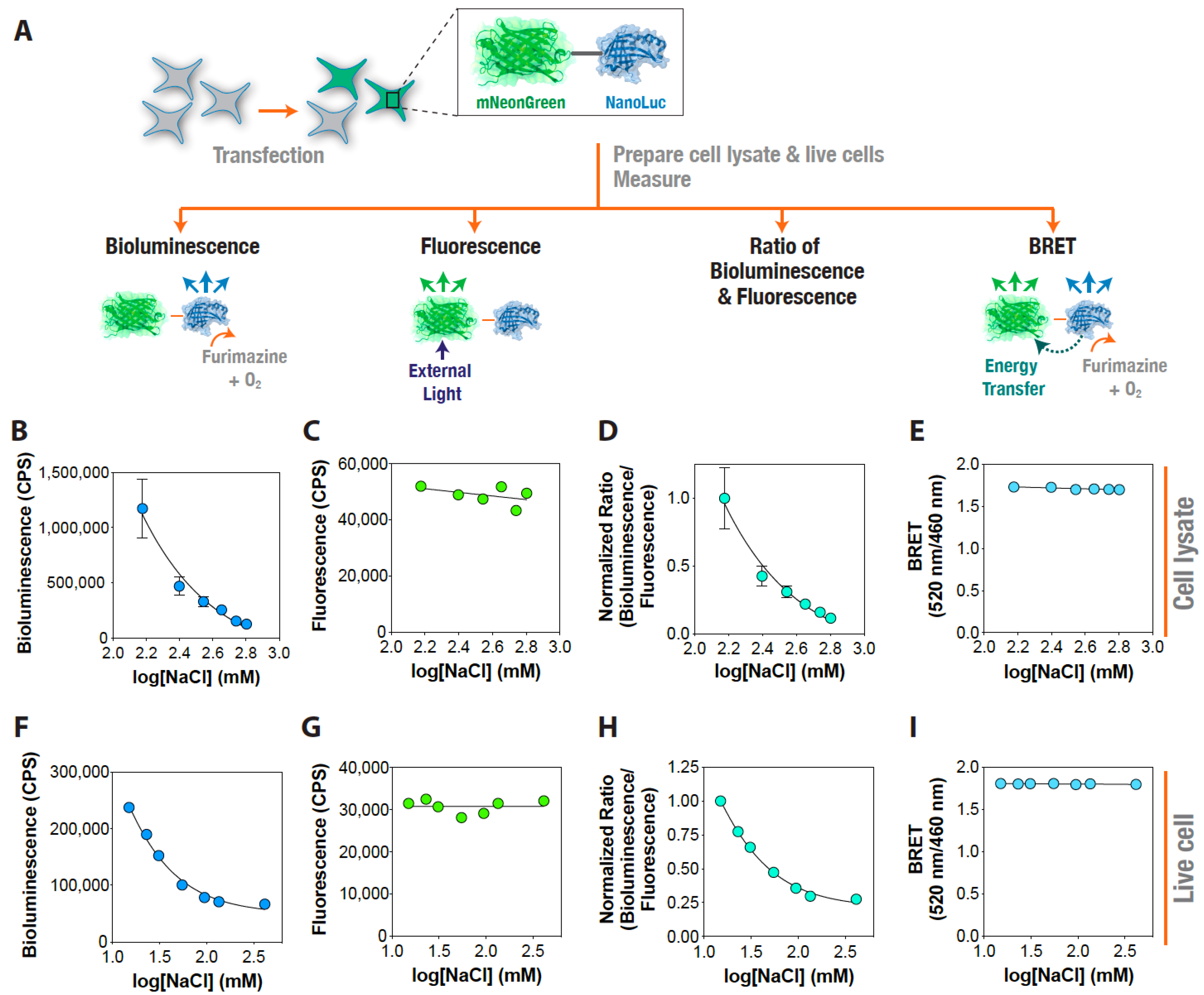
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar] [CrossRef] [Green Version]
- Warshel, A.; Russell, S.T. Calculations of electrostatic interactions in biological systems and in solutions. Q. Rev. Biophys. 2009, 17, 283–422. [Google Scholar] [CrossRef]
- Altamash, T.; Coronas, A. Interactions in l-histidine/l-glutamic acid/l-tryptophan/glycylglycine + 2 mol L−1 aqueous KCl/KNO3 systems at different temperatures: An isothermal compressibility study. Thermochim. Acta 2012, 543, 313–317. [Google Scholar] [CrossRef]
- Andre, A.A.M.; Spruijt, E. Liquid-Liquid Phase Separation in Crowded Environments. Int. J. Mol. Sci. 2020, 21, 5908. [Google Scholar] [CrossRef]
- König, I.; Zarrine-Afsar, A.; Aznauryan, M.; Soranno, A.; Wunderlich, B.; Dingfelder, F.; Stüber, J.C.; Plückthun, A.; Nettels, D.; Schuler, B. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 2015, 12, 773–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, K.H.; Badireddy, S.; Rajendran, A.; Anand, G.S.; Visweswariah, S.S. Cyclic nucleotide binding and structural changes in the isolated GAF domain of Anabaena adenylyl cyclase, CyaB2. PeerJ 2015, 3, e882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, K.H.; Visweswariah, S.S. Buffer NaCl concentration regulates Renilla luciferase activity and ligand-induced conformational changes in the BRET-based PDE5 sensor. Matters 2017. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.H. Allosteric regulation of proteins. Resonance 2017, 22, 37–50. [Google Scholar] [CrossRef]
- Biswas, K.H.; Visweswariah, S.S. Distinct allostery induced in the cyclic GMP-binding, cyclic GMP-specific phosphodiesterase (PDE5) by cyclic GMP, sildenafil, and metal ions. J. Biol. Chem. 2011, 286, 8545–8554. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.H.; Sopory, S.; Visweswariah, S.S. The GAF domain of the cGMP-binding, cGMP-specific phosphodiesterase (PDE5) is a sensor and a sink for cGMP. Biochemistry 2008, 47, 3534–3543. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Nidetzky, B. The Microenvironment in Immobilized Enzymes: Methods of Characterization and Its Role in Determining Enzyme Performance. Molecules 2019, 24, 3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikfarjam, S.; Jouravleva, E.V.; Anisimov, M.A.; Woehl, T.J. Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions. Biomolecules 2020, 10, 1262. [Google Scholar] [CrossRef]
- Marek, P.J.; Patsalo, V.; Green, D.F.; Raleigh, D.P. Ionic Strength Effects on Amyloid Formation by Amylin Are a Complicated Interplay among Debye Screening, Ion Selectivity, and Hofmeister Effects. Biochemistry 2012, 51, 8478–8490. [Google Scholar] [CrossRef] [PubMed]
- Mari, E.; Ricci, C.; Pieraccini, S.; Spinozzi, F.; Mariani, P.; Ortore, M.G. Trehalose Effect on The Aggregation of Model Proteins into Amyloid Fibrils. Life 2020, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Mikalauskaite, K.; Ziaunys, M.; Sneideris, T.; Smirnovas, V. Effect of Ionic Strength on Thioflavin-T Affinity to Amyloid Fibrils and Its Fluorescence Intensity. Int. J. Mol. Sci. 2020, 21, 8916. [Google Scholar] [CrossRef] [PubMed]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.-H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhofer, W.; Bartels, H.; Fraek, M.-L.; Beck, F.-X. Relationship between intracellular ionic strength and expression of tonicity-responsive genes in rat papillary collecting duct cells. J. Physiol. 2002, 543, 147. [Google Scholar] [CrossRef]
- Higgins, C.F.; Cairney, J.; Stirling, D.A.; Sutherland, L.; Booth, I.R. Osmotic regulation of gene expression: Ionic strength as an intracellular signal? Trends Biochem. Sci. 1987, 12, 339–344. [Google Scholar] [CrossRef]
- Syeda, R.; Qiu, Z.; Dubin, A.E.; Murthy, S.E.; Florendo, M.N.; Mason, D.E.; Mathur, J.; Cahalan, S.M.; Peters, E.C.; Montal, M. LRRC8 proteins form volume-regulated anion channels that sense ionic strength. Cell 2016, 164, 499. [Google Scholar] [CrossRef] [Green Version]
- Sabapathy, T.; Deplazes, E.; Mancera, R.L. Revisiting the Interaction of Melittin with Phospholipid Bilayers: The Effects of Concentration and Ionic Strength. Int. J. Mol. Sci. 2020, 21, 746. [Google Scholar] [CrossRef] [Green Version]
- Machan, R.; Hof, M. Recent developments in fluorescence correlation spectroscopy for diffusion measurements in planar lipid membranes. Int. J. Mol. Sci. 2010, 11, 427–457. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, E.; Lambert, I.; Pedersen, S. Physiology of cell volume regulation in vertebrates. Physiol. Rev. 2009, 89, 193. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Poolman, B.; Boersma, A.J. Ionic Strength Sensing in Living Cells. ACS Chem. Biol. 2017, 12, 2510. [Google Scholar] [CrossRef] [Green Version]
- Moussa, R.; Baierl, A.; Steffen, V.; Kubitzki, T.; Wiechert, W.; Pohl, M. An evaluation of genetically encoded FRET-based biosensors for quantitative metabolite analyses in vivo. J. Biotechnol. 2014, 191, 250. [Google Scholar] [CrossRef] [PubMed]
- Biemans-Oldehinkel, E.; Mahmood, N.A.; Poolman, B. A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. USA 2006, 103, 10624. [Google Scholar] [CrossRef] [Green Version]
- Cortese, J.; Voglino, A.; Hackenbrock, C. Ionic strength of the intermembrane space of intact mitochondria as estimated with fluorescein-BSA delivered by low pH fusion. J. Cell Biol. 1991, 113, 1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leavesley, S.; Rich, T. Overcoming limitations of FRET measurements. Cytom. Part A J. Int. Soc. Anal. Cytol. 2016, 89, 325. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848. [Google Scholar] [CrossRef] [PubMed]
- Sureda-Vives, M.; Sarkisyan, K.S. Bioluminescence-Driven Optogenetics. Life 2020, 10, 318. [Google Scholar] [CrossRef]
- Endo, M.; Ozawa, T. Advanced Bioluminescence System for In Vivo Imaging with Brighter and Red-Shifted Light Emission. Int. J. Mol. Sci. 2020, 21, 6538. [Google Scholar] [CrossRef]
- Samanta, A.; Medintz, I.L. Bioluminescence-Based Energy Transfer Using Semiconductor Quantum Dots as Acceptors. Sensors 2020, 20, 2909. [Google Scholar] [CrossRef] [PubMed]
- Dale, N.C.; Johnstone, E.K.M.; White, C.W.; Pfleger, K.D.G. NanoBRET: The Bright Future of Proximity-Based Assays. Front. Bioeng. Biotechnol. 2019, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Wu, Q.P.; Xu, T.; Liu, Y.L.; Xu, Z.G.; Zhang, S.F.; Guo, Z.Y. Quick preparation of nanoluciferase-based tracers for novel bioluminescent receptor-binding assays of protein hormones: Using erythropoietin as a model. J. Photochem. Photobiol. B 2015, 153, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, L.; Cheng, X.; Ge, X.; Wang, P. An ultrasensitive system for measuring the USPs and OTULIN activity using Nanoluc as a reporter. Biochem. Biophys. Res. Commun. 2014, 455, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, G.; Shao, X.X.; Liu, Y.L.; Guo, Z.Y. Quantitative measurement of cell membrane receptor internalization by the nanoluciferase reporter: Using the G protein-coupled receptor RXFP3 as a model. Biochim. Biophys. Acta 2015, 1848, 688–694. [Google Scholar] [CrossRef] [Green Version]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Lackner, D.H.; Carre, A.; Guzzardo, P.M.; Banning, C.; Mangena, R.; Henley, T.; Oberndorfer, S.; Gapp, B.V.; Nijman, S.M.B.; Brummelkamp, T.R.; et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015, 6, 10237. [Google Scholar] [CrossRef]
- Gaspar, N.; Zambito, G.; Dautzenberg, I.J.C.; Cramer, S.J.; Hoeben, R.C.; Lowik, C.; Walker, J.R.; Kirkland, T.A.; Smith, T.P.; van Weerden, W.M.; et al. NanoBiT System and Hydrofurimazine for Optimized Detection of Viral Infection in Mice-A Novel in Vivo Imaging Platform. Int. J. Mol. Sci. 2020, 21, 5863. [Google Scholar] [CrossRef]
- El Khamlichi, C.; Reverchon-Assadi, F.; Hervouet-Coste, N.; Blot, L.; Reiter, E.; Morisset-Lopez, S. Bioluminescence Resonance Energy Transfer as a Method to Study Protein-Protein Interactions: Application to G Protein Coupled Receptor Biology. Molecules 2019, 24, 537. [Google Scholar] [CrossRef] [Green Version]
- Arts, R.; Ludwig, S.K.J.; van Gerven, B.C.B.; Estirado, E.M.; Milroy, L.G.; Merkx, M. Semisynthetic Bioluminescent Sensor Proteins for Direct Detection of Antibodies and Small Molecules in Solution. ACS Sens. 2017, 2, 1730–1736. [Google Scholar] [CrossRef]
- Mie, M.; Hirashima, R.; Mashimo, Y.; Kobatake, E. Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor. Appl. Sci. 2020, 10, 7646. [Google Scholar] [CrossRef]
- Min, S.H.; French, A.R.; Trull, K.J.; Tat, K.; Varney, S.A.; Tantama, M. Ratiometric BRET Measurements of ATP with a Genetically-Encoded Luminescent Sensor. Sensors 2019, 19, 3502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mocking, T.A.M.B.; Buzink, M.C.M.L.; Leurs, R.; Vischer, H.F. Bioluminescence Resonance Energy Transfer Based G Protein-Activation Assay to Probe Duration of Antagonism at the Histamine H3 Receptor. Int. J. Mol. Sci. 2019, 20, 3724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimmer, T.; Schroeter, E.; Lorenz, B.; Stieger, K. Detection of the Vascular Endothelial Growth Factor with a Novel Bioluminescence Resonance Energy Transfer Pair Using a Two-Component System. Sensors 2017, 17, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.N.; Ishida, R.; Hattori, M.; Matsuda, T.; Nagai, T. Bioluminescent Ratiometric Indicator for Analysis of Water Hardness in Household Water. Sensors 2020, 20, 3164. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, Z.J. NanoLuc Bioluminescence-Driven Photodynamic Activation of Cholecystokinin 1 Receptor with Genetically-Encoded Protein Photosensitizer MiniSOG. Int. J. Mol. Sci. 2020, 21, 3763. [Google Scholar] [CrossRef]
- Belarbi, E.; Legros, V.; Basset, J.; Despres, P.; Roques, P.; Choumet, V. Bioluminescent Ross River Virus Allows Live Monitoring of Acute and Long-Term Alphaviral Infection by In Vivo Imaging. Viruses 2019, 11, 584. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Yang, X.; Wang, Y.; Shen, X. In Vivo Analysis of Protein-Protein Interactions with Bioluminescence Resonance Energy Transfer (BRET): Progress and Prospects. Int. J. Mol. Sci. 2016, 17, 1704. [Google Scholar] [CrossRef] [Green Version]
- Nath, N.; Flemming, R.; Godat, B.; Urh, M. Development of NanoLuc bridging immunoassay for detection of anti-drug antibodies. J. Immunol. Methods 2017, 450, 17–26. [Google Scholar] [CrossRef]
- Boute, N.; Lowe, P.; Berger, S.; Malissard, M.; Robert, A.; Tesar, M. NanoLuc Luciferase–A Multifunctional Tool for High Throughput Antibody Screening. Front. Pharmacol. 2016, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, J.V.; Bernardi, R.C.; Rudack, T.; Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Schulten, K. QwikMD—Integrative Molecular Dynamics Toolkit for Novices and Experts. Sci. Rep. 2016, 6, 26536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- den Hamer, A.; Dierickx, P.; Arts, R.; de Vries, J.; Brunsveld, L.; Merkx, M. Bright Bioluminescent BRET Sensor Proteins for Measuring Intracellular Caspase Activity. ACS Sens. 2017, 2, 729–734. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.H.; Zhongwen, C.; Dubey, A.K.; Oh, D.; Groves, J.T. Multicomponent Supported Membrane Microarray for Monitoring Spatially Resolved Cellular Signaling Reactions. Adv. Biosyst. 2018, 2, 1800015. [Google Scholar] [CrossRef]
- Biswas, K.H.; Hartman, K.L.; Zaidel-Bar, R.; Groves, J.T. Sustained alpha-catenin Activation at E-cadherin Junctions in the Absence of Mechanical Force. Biophys. J. 2016, 111, 1044–1052. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.H.; Hartman, K.L.; Yu, C.H.; Harrison, O.J.; Song, H.; Smith, A.W.; Huang, W.Y.; Lin, W.C.; Guo, Z.; Padmanabhan, A.; et al. E-cadherin junction formation involves an active kinetic nucleation process. Proc. Natl. Acad. Sci. USA 2015, 112, 10932–10937. [Google Scholar] [CrossRef] [Green Version]
- Yeh, H.-W.; Karmach, O.; Ji, A.; Carter, D.; Martins-Green, M.M.; Ai, H.-W. Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 2017, 14, 971. [Google Scholar] [CrossRef]
- Klein, C.; Sunahara, R.; Hudson, T.; Heyduk, T.; Howlett, A. Zinc inhibition of cAMP signaling. J. Biol. Chem. 2002, 277, 11859. [Google Scholar] [CrossRef] [Green Version]
- Loening, A.M.; Fenn, T.D.; Gambhir, S.S. Crystal Structures of the Luciferase and Green Fluorescent Protein from Renilla reniformis. J. Mol. Biol. 2007, 374, 1017. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altamash, T.; Ahmed, W.; Rasool, S.; Biswas, K.H. Intracellular Ionic Strength Sensing Using NanoLuc. Int. J. Mol. Sci. 2021, 22, 677. https://doi.org/10.3390/ijms22020677
Altamash T, Ahmed W, Rasool S, Biswas KH. Intracellular Ionic Strength Sensing Using NanoLuc. International Journal of Molecular Sciences. 2021; 22(2):677. https://doi.org/10.3390/ijms22020677
Chicago/Turabian StyleAltamash, Tausif, Wesam Ahmed, Saad Rasool, and Kabir H. Biswas. 2021. "Intracellular Ionic Strength Sensing Using NanoLuc" International Journal of Molecular Sciences 22, no. 2: 677. https://doi.org/10.3390/ijms22020677
APA StyleAltamash, T., Ahmed, W., Rasool, S., & Biswas, K. H. (2021). Intracellular Ionic Strength Sensing Using NanoLuc. International Journal of Molecular Sciences, 22(2), 677. https://doi.org/10.3390/ijms22020677




