Differential DNA Methylation in Prostate Tumors from Puerto Rican Men
Abstract
:1. Introduction
2. Results
2.1. Clinicopathological Characteristics of Study Group
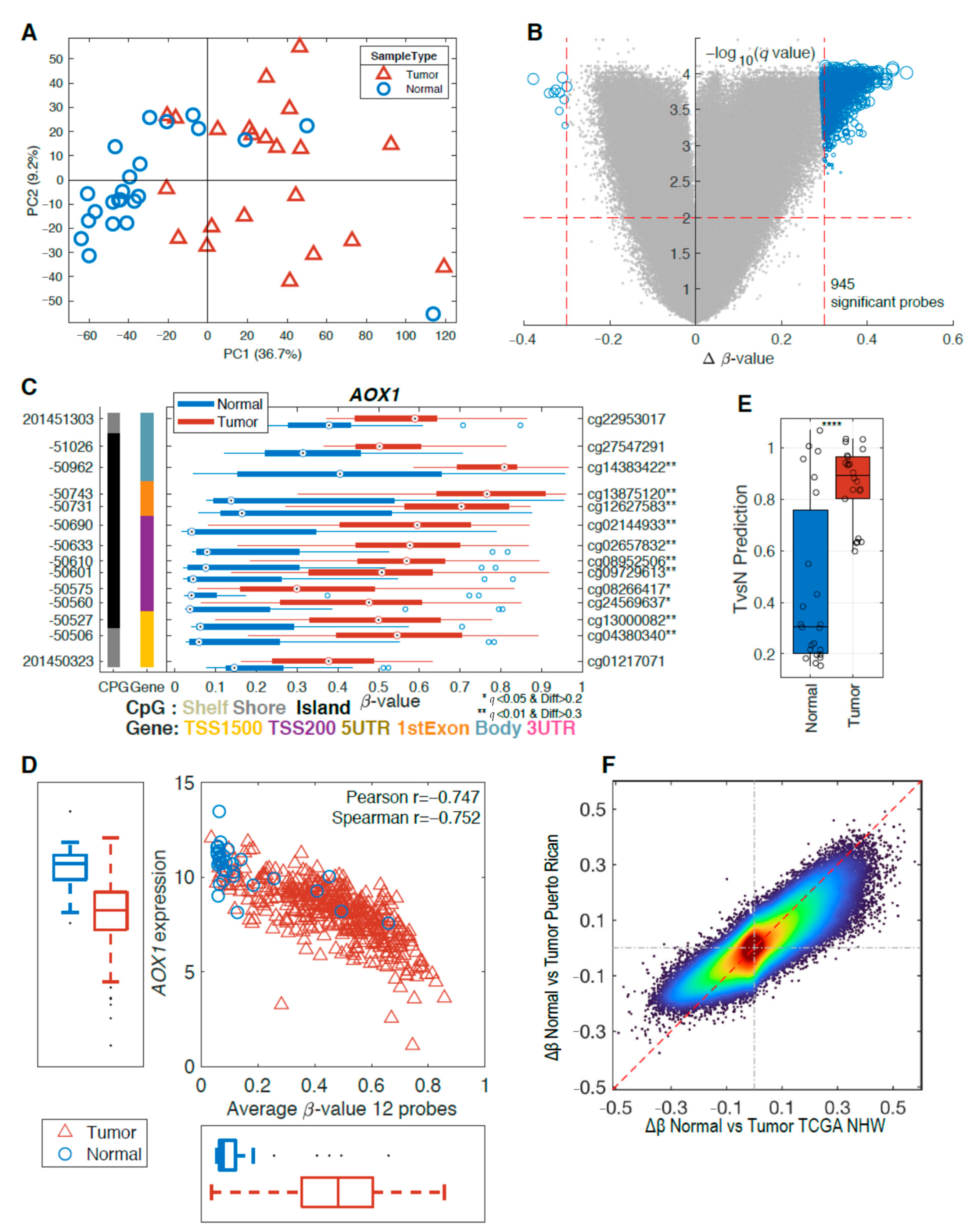
2.2. Differences in Methylation Levels between Tumors and Adjacent Normal Tissue
2.3. Differentially Methylated Genes Associated with Aggressive Prostate Cancer
2.4. Methylation Analysis of DNA Repair Genes Associated with Aggressive Prostate Cancer
2.5. Ancestry Analysis
3. Discussion
4. Materials and Methods
4.1. Human Subjects, Sample Selection, and Specimen Acquisition
4.2. DNA Methylation
4.2.1. Illumina Methylation 850K Data Filtering and Genome-Wide Analysis Plan
4.2.2. DNA Extraction
4.2.3. Quality Control (QC) and Normalization for Methylation Data
4.2.4. Selection of Differentially Methylated Regions (DMRs)
4.2.5. TCGA Data
4.3. Ancestry Analysis
4.3.1. Genotyping
4.3.2. Quality Control of Ancestry Data
4.3.3. Ancestry Estimates
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCa | Prostate cancer |
| DRC | DNA repair capacity |
| NER | Nucleotide excision repair |
| IRB | Institutional Review Board |
| DMR | Differentially methylated region |
| PR | Puerto Rico |
| H/L | Hispanic Latino |
| AA | African American |
| NHW | Non-Hispanic White |
| PCA | Principal component analysis |
| AIMS | Ancestry informative markers |
| QA | Quality assurance |
| QC | Quality control |
| FFPE | Formalin-fixed paraffin-embedded |
| SNPs | Single-nucleotide polymorphisms |
| TCGA | The Cancer Genome Atlas |
| FDR | False discovery rate |
| PCR | Polymerase chain reaction |
| PHSU | Ponce Health Sciences University |
| PRI | Ponce Research Institute |
| PRBB | Puerto Rico Biobank |
| HR | Homologous recombination |
| PARP1 | Poly (ADP-Ribose) polymerase-1 |
| BRCA1 | Breast cancer type 1 susceptibility protein |
| BRCA2 | Breast cancer type 2 susceptibility protein |
| DSBs | DNA double-strand breaks |
| AOX1 | Aldehyde Oxidase 1 |
| RREB1 | Ras Responsive Element Binding Protein 1 |
| FAM71F2 | Family with Sequence Similarity 71 F2 |
| JMJD1C | Jumonji Domain Containing 1C |
| COL5A3 | Collagen Type V Alpha 3 Chain |
| RAE1 | Ribonucleic Acid Export 1 |
| GABRQ | Gamma-Aminobutyric Acid Type A Receptor Subunit Theta |
| COL9A2 | Collagen Type IX Alpha 2 Chain |
| FAM179A | TOG Array Regulator of Axonemal Microtubules 2 |
| SLC17A2 | Solute Carrier Family 17 Member 2 |
| PDE10 | Phosphodiesterase 10 |
| PLEKHS1 | Pleckstrin Homology Domain Containing S1 |
| TNNI2 | Troponin I2, Fast Skeletal Type |
| OR51A4 | Olfactory Receptor Family 51 Subfamily A Member 4 |
| RNF169 | Ring Finger Protein 169 |
| SPNS2 | Sphingolipid Transporter 2 |
| ADAMTSL5 | ADAMTS Like 5 |
| CYP4F12 | Cytochrome P450 Family 4 Subfamily F Member 12 |
| MDC1 | Mediator of DNA Damage Checkpoint 1 |
| PARP | Poly(ADP-Ribose) Polymerase |
| BRCA1 | BRCA1 DNA Repair Associated |
| BRCA2 | BRCA2 DNA Repair Associated |
References
- NCI. Cancer Stat Facts: Prostate Cancer; National Cancer Institute: Surveillance, Epidemiology, and End Results Program; Department of Health and Human Services: Bethesda, MD USA, 2018. [Google Scholar]
- American Cancer Socitey. Cancer Facts & Figures 2020; American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Chinea, F.M.; Patel, V.N.; Kwon, D.; Lamichhane, N.; Lopez, C.; Punnen, S.; Kobetz, E.N.; Abramowitz, M.C.; Pollack, A. Ethnic heterogeneity and prostate cancer mortality in Hispanic/Latino men: A population-based study. Oncotarget 2017, 8, 69709–69721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tortolero-Luna, G.; Zavala-Zegarra, D.; Pérez-Ríos, N.; Torres-Cintrón, C.R.; Ortíz-Ortíz, K.J.; Traverso-Ortíz, M.; Román-Ruiz, Y.; Veguilla-Rosario, I.; Vázquez-Cubano, N.; Merced-Vélez, M.F.; et al. Cancer in Puerto Rico, 2006–2010; Puerto Rico Central Cancer Registry: San Juan, PR, USA, 2013. [Google Scholar]
- Ukimura, O.; Coleman, J.A.; de la Taille, A.; Emberton, M.; Epstein, J.I.; Freedland, S.J.; Giannarini, G.; Kibel, A.S.; Montironi, R.; Ploussard, G.; et al. Contemporary role of systematic prostate biopsies: Indications, techniques, and implications for patient care. Eur. Urol. 2013, 63, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Graif, T.; Loeb, S.; Roehl, K.A.; Gashti, S.N.; Griffin, C.; Yu, X.; Catalona, W.J. Under diagnosis and over diagnosis of prostate cancer. J. Urol. 2007, 178, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.; Ward, K.C.; Osunkoya, A.O.; Datta, M.W.; Luthringer, D.; Young, A.N.; Marks, K.; Cohen, V.; Kennedy, J.C.; Haber, M.J.; et al. Frequency and determinants of disagreement and error in gleason scores: A population-based study of prostate cancer. Prostate 2012, 72, 1389–1398. [Google Scholar] [CrossRef] [Green Version]
- Nikas, J.B.; Nikas, E.G. Genome-Wide DNA Methylation Model for the Diagnosis of Prostate Cancer. ACS Omega 2019, 4, 14895–14901. [Google Scholar] [CrossRef] [Green Version]
- Kurfurstova, D.; Bartkova, J.; Vrtel, R.; Mickova, A.; Burdova, A.; Majera, D.; Mistrik, M.; Kral, M.; Santer, F.R.; Bouchal, J.; et al. DNA damage signalling barrier, oxidative stress and treatment-relevant DNA repair factor alterations during progression of human prostate cancer. Mol. Oncol. 2016, 10, 879–894. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Ambrosone, C.B.; Lee, J.; Sellers, T.A.; Pow-Sang, J.; Park, J.Y. Association between polymorphisms in the DNA repair genes XRCC1 and APE1, and the risk of prostate cancer in white and black Americans. J. Urol. 2006, 175, 108–112; discussion 112. [Google Scholar] [CrossRef]
- Park, J.Y.; Huang, Y.; Sellers, T.A. Single nucleotide polymorphisms in DNA repair genes and prostate cancer risk. Methods Mol. Biol. 2009, 471, 361–385. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Wu, Y.P.; Ke, Z.B.; Liang, Y.C.; Cai, H.; Su, W.T.; Tao, X.; Chen, S.H.; Zheng, Q.S.; Wei, Y.; et al. Identification of key DNA methylation-driven genes in prostate adenocarcinoma: An integrative analysis of TCGA methylation data. J. Transl. Med. 2019, 17, 311. [Google Scholar] [CrossRef]
- Pecorino, L. Molecular Biology of Cancer-Mechanisms, Targets and Therapeutics, 4th ed.; Oxford University Press: Oxford UK, 2016. [Google Scholar]
- Carleton, N.M.; Zhu, G.; Gorbounov, M.; Miller, M.C.; Pienta, K.J.; Resar, L.M.S.; Veltri, R.W. PBOV1 as a potential biomarker for more advanced prostate cancer based on protein and digital histomorphometric analysis. Prostate 2018, 78, 547–559. [Google Scholar] [CrossRef]
- Komura, K.; Sweeney, C.J.; Inamoto, T.; Ibuki, N.; Azuma, H.; Kantoff, P.W. Current treatment strategies for advanced prostate cancer. Int. J. Urol. 2018, 25, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giudice, A.; Montella, M.; Boccellino, M.; Crispo, A.; D’Arena, G.; Bimonte, S.; Facchini, G.; Ciliberto, G.; Botti, G.; Quagliuolo, L.; et al. Epigenetic Changes Induced by Green Tea Catechins are Associated with Prostate Cancer. Curr. Mol. Med. 2017, 17, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y. Promoter hypermethylation in prostate cancer. Cancer Control 2010, 17, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Park, J.Y. DNA methylation in promoter region as biomarkers in prostate cancer. Methods Mol. Biol. 2012, 863, 67–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaney, J.M.; Wang, S.; Funda, S.; Long, J.; Taghipour, D.J.; Tbaishat, R.; Furbert-Harris, P.; Ittmann, M.; Kwabi-Addo, B. Identification of novel DNA-methylated genes that correlate with human prostate cancer and high-grade prostatic intraepithelial neoplasia. Prostate Cancer Prostatic Dis. 2013, 16, 292–300. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.W.; Kim, S.T.; Turner, A.R.; Young, T.; Smith, S.; Liu, W.; Lindberg, J.; Egevad, L.; Gronberg, H.; Isaacs, W.B.; et al. Identification of new differentially methylated genes that have potential functional consequences in prostate cancer. PLoS ONE 2012, 7, e48455. [Google Scholar] [CrossRef] [Green Version]
- Geybels, M.S.; Zhao, S.; Wong, C.J.; Bibikova, M.; Klotzle, B.; Wu, M.; Ostrander, E.A.; Fan, J.B.; Feng, Z.; Stanford, J.L. Epigenomic profiling of DNA methylation in paired prostate cancer versus adjacent benign tissue. Prostate 2015, 75, 1941–1950. [Google Scholar] [CrossRef] [Green Version]
- Apprey, V.; Wang, S.; Tang, W.; Kittles, R.A.; Southerland, W.M.; Ittmann, M.; Kwabi-Addo, B. Association of Genetic Ancestry With DNA Methylation Changes in Prostate Cancer Disparity. Anticancer Res. 2019, 39, 5861–5866. [Google Scholar] [CrossRef]
- Strand, S.H.; Orntoft, T.F.; Sorensen, K.D. Prognostic DNA methylation markers for prostate cancer. Int. J. Mol. Sci. 2014, 15, 16544–16576. [Google Scholar] [CrossRef] [Green Version]
- Ruf, C.G.; Linbecker, M.; Port, M.; Riecke, A.; Schmelz, H.U.; Wagner, W.; Meineke, V.; Abend, M. Predicting metastasized seminoma using gene expression. BJU Int. 2012, 110, E14–E20. [Google Scholar] [CrossRef]
- An, L.; Dong, C.; Li, J.; Chen, J.; Yuan, J.; Huang, J.; Chan, K.M.; Yu, C.-h.; Huen, M.S.Y. RNF169 limits 53BP1 deposition at DSBs to stimulate single-strand annealing repair. Proc. Natl. Acad. Sci. USA 2018, 115, E8286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Hao, X.; Song, Y. Identification of the Key MicroRNAs and the miRNA-mRNA Regulatory Pathways in Prostate Cancer by Bioinformatics Methods. Biomed. Res. Int. 2018, 2018, 6204128. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, N.K.; Ferdinand, A.S.; Mukhopadhyay, L.; Cinar, B.; Lutchman, M.; Richie, J.P.; Freeman, M.R.; Liu, B.C. Unraveling androgen receptor interactomes by an array-based method: Discovery of proto-oncoprotein c-Rel as a negative regulator of androgen receptor. Exp. Cell Res. 2006, 312, 3782–3795. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, N.K.; Cinar, B.; Mukhopadhyay, L.; Lutchman, M.; Ferdinand, A.S.; Kim, J.; Chung, L.W.; Adam, R.M.; Ray, S.K.; Leiter, A.B.; et al. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: Implications for the role of the Ras pathway in enhancing androgenic signaling in prostate cancer. Mol. Endocrinol. 2007, 21, 2056–2070. [Google Scholar] [CrossRef] [Green Version]
- Mukund, K.; Syulyukina, N.; Ramamoorthy, S.; Subramaniam, S. Right and left-sided colon cancers—Specificity of molecular mechanisms in tumorigenesis and progression. BMC Cancer 2020, 20, 317. [Google Scholar] [CrossRef] [Green Version]
- Eun, H.S.; Cho, S.Y.; Lee, B.S.; Seong, I.O.; Kim, K.H. Profiling cytochrome P450 family 4 gene expression in human hepatocellular carcinoma. Mol. Med. Rep. 2018, 18, 4865–4876. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.L.; Wang, L.; Cao, Z.Y.; Wang, J.; Jing, M.Z.; Xia, Z.C.; Ao, F.; Ye, L.B.; Liu, S.; Zhu, Y. Inducible CYP4F12 enhances Hepatitis C virus infection via association with viral nonstructural protein 5B. Biochem. Biophys. Res. Commun. 2016, 471, 95–102. [Google Scholar] [CrossRef]
- Huang, W.; Zhao, C.; Zhong, H.; Zhang, S.; Xia, Y.; Cai, Z. Bisphenol S induced epigenetic and transcriptional changes in human breast cancer cell line MCF-7. Environ. Pollut. 2019, 246, 697–703. [Google Scholar] [CrossRef]
- Abdullah, M.; Choo, C.W.; Alias, H.; Abdul Rahman, E.J.; Mohd Ibrahim, H.; Jamal, R.; Hussin, N.H. ADAMTSL5 and CDH11: Putative epigenetic markers for therapeutic resistance in acute lymphoblastic leukemia. Hematology 2017, 22, 386–391. [Google Scholar] [CrossRef] [Green Version]
- Li, T.Q.; Feng, C.Q.; Zou, Y.G.; Shi, R.; Liang, S.; Mao, X.M. Literature-mining and bioinformatic analysis of androgen-independent prostate cancer-specific genes. Zhonghua Nan Ke Xue 2009, 15, 1102–1107. [Google Scholar]
- Fang, L.; Hou, J.; Cao, Y.; Shan, J.J.; Zhao, J. Spinster homolog 2 in cancers, its functions and mechanisms. Cell. Signal. 2020, 109821. [Google Scholar] [CrossRef]
- Brizuela, L.; Martin, C.; Jeannot, P.; Ader, I.; Gstalder, C.; Andrieu, G.; Bocquet, M.; Laffosse, J.M.; Gomez-Brouchet, A.; Malavaud, B.; et al. Osteoblast-derived sphingosine 1-phosphate to induce proliferation and confer resistance to therapeutics to bone metastasis-derived prostate cancer cells. Mol. Oncol. 2014, 8, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.; Dasgupta, S.; Jiang, X.; Zhao, X.; Zhu, G.; He, Q.; Dinkins, M.; Bieberich, E.; Wang, G. Critical role of Spns2, a sphingosine-1-phosphate transporter, in lung cancer cell survival and migration. PLoS ONE 2014, 9, e110119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Qian, T.; Cheng, Z.; Zeng, T.; Si, C.; Liu, C.; Deng, C.; Ye, X.; Liu, Y.; Cui, L.; et al. Prognostic significance of Spinster homolog gene family in acute myeloid leukemia. J. Cancer 2020, 11, 4581–4588. [Google Scholar] [CrossRef] [PubMed]
- Sawaki, K.; Kanda, M.; Miwa, T.; Umeda, S.; Tanaka, H.; Tanaka, C.; Kobayashi, D.; Suenaga, M.; Hattori, N.; Hayashi, M.; et al. Troponin I2 as a Specific Biomarker for Prediction of Peritoneal Metastasis in Gastric Cancer. Ann. Surg. Oncol. 2018, 25, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Zengin, T.; Onal-Suzek, T. Analysis of genomic and transcriptomic variations as prognostic signature for lung adenocarcinoma. BMC Bioinform. 2020, 21, 368. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, Y.; Yan, L.L.; An, R.; Wang, X.Y.; Wang, H.Y. Exploring DNA Methylation Profiles Altered in Cryptogenic Hepatocellular Carcinomas by High-Throughput Targeted DNA Methylation Sequencing: A Preliminary Study for Cryptogenic Hepatocellular Carcinoma. OncoTargets Ther. 2020, 13, 9901–9916. [Google Scholar] [CrossRef]
- Xing, X.; Mu, N.; Yuan, X.; Wang, N.; Juhlin, C.C.; Straat, K.; Larsson, C.; Xu, D. PLEKHS1 Over-Expression is Associated with Metastases and Poor Outcomes in Papillary Thyroid Carcinoma. Cancers 2020, 12, 2133. [Google Scholar] [CrossRef]
- Yan, J.; Zhou, X.; Pan, D. A case of one lung adenocarcinoma patient harboring a novel FAM179A-ALK (F1, A19) rearrangement responding to lorlatinib treatment. Lung Cancer 2020, 147, 26–29. [Google Scholar] [CrossRef]
- Lee, D.; Ha, M.; Hong, C.M.; Kim, J.; Park, S.M.; Park, D.; Sohn, D.H.; Shin, H.J.; Yu, H.S.; Kim, C.D.; et al. GABRQ expression is a potential prognostic marker for patients with clear cell renal cell carcinoma. Oncol. Lett. 2019, 18, 5731–5738. [Google Scholar] [CrossRef]
- Li, Y.H.; Liu, Y.; Li, Y.D.; Liu, Y.H.; Li, F.; Ju, Q.; Xie, P.L.; Li, G.C. GABA stimulates human hepatocellular carcinoma growth through overexpressed GABAA receptor theta subunit. World J. Gastroenterol. WJG 2012, 18, 2704–2711. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.A.; Ayyagari, R.R.; Wheeler, G.L.; Weiss, R.M. Regulation of cyclic nucleotides in the urinary tract. J. Smooth Muscle Res. 2005, 41, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, K.; Maehara, S.; Yuge, N.; Ishikawa, M.; Miyazaki, Y.; Naba, H.; Kato, Y.; Nakao, K. Pharmacological characterization of a novel potent, selective, and orally active phosphodiesterase 10A inhibitor, PDM-042 [(E)-4-(2-(2-(5,8-dimethyl-[1,2,4]triazolo[1,5-a]pyrazin-2-yl)vinyl)-6-(pyrrolidi n-1-yl)pyrimidin-4-yl)morpholine] in rats: Potential for the treatment of schizophrenia. Pharmacol. Res. Perspect. 2016, 4, e00241. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lee, K.; Xi, Y.; Zhu, B.; Gary, B.D.; Ramirez-Alcantara, V.; Gurpinar, E.; Canzoneri, J.C.; Fajardo, A.; Sigler, S.; et al. Phosphodiesterase 10A: A novel target for selective inhibition of colon tumor cell growth and beta-catenin-dependent TCF transcriptional activity. Oncogene 2015, 34, 1499–1509. [Google Scholar] [CrossRef] [Green Version]
- Hennenberg, M.; Schott, M.; Kan, A.; Keller, P.; Tamalunas, A.; Ciotkowska, A.; Rutz, B.; Wang, Y.; Strittmatter, F.; Herlemann, A.; et al. Inhibition of Adrenergic and Non-Adrenergic Smooth Muscle Contraction in the Human Prostate by the Phosphodiesterase 10-Selective Inhibitor TC-E 5005. Prostate 2016, 76, 1364–1374. [Google Scholar] [CrossRef]
- Guard, S.E.; Poss, Z.C.; Ebmeier, C.C.; Pagratis, M.; Simpson, H.; Taatjes, D.J.; Old, W.M. The nuclear interactome of DYRK1A reveals a functional role in DNA damage repair. Sci. Rep. 2019, 9, 6539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, V.R.; Ananthapadmanabhan, V.; Swanson, S.; Saini, S.; Sesay, F.; Yakovlev, V.; Florens, L.; DeCaprio, J.A.; Washburn, M.P.; Dozmorov, M.; et al. DYRK1A regulates the recruitment of 53BP1 to the sites of DNA damage in part through interaction with RNF169. Cell Cycle 2019, 18, 531–551. [Google Scholar] [CrossRef] [Green Version]
- Roewenstrunk, J.; Di Vona, C.; Chen, J.; Borras, E.; Dong, C.; Arato, K.; Sabido, E.; Huen, M.S.Y.; de la Luna, S. A comprehensive proteomics-based interaction screen that links DYRK1A to RNF169 and to the DNA damage response. Sci. Rep. 2019, 9, 6014. [Google Scholar] [CrossRef]
- Laham, A.J.; Saber-Ayad, M.; El-Awady, R. DYRK1A: A down syndrome-related dual protein kinase with a versatile role in tumorigenesis. Cell. Mol. Life Sci. 2020, 1–7. [Google Scholar] [CrossRef]
- Frank, S.; Nelson, P.; Vasioukhin, V. Recent advances in prostate cancer research: Large-scale genomic analyses reveal novel driver mutations and DNA repair defects. F1000Research 2018, 2. [Google Scholar] [CrossRef]
- Dhawan, M.; Ryan, C.J.; Ashworth, A. DNA Repair Deficiency Is Common in Advanced Prostate Cancer: New Therapeutic Opportunities. Oncologist 2016, 21, 940–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.-P.; Triche, T.J., Jr.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2016, 33, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Triche, T.J., Jr.; Weisenberger, D.J.; Van Den Berg, D.; Laird, P.W.; Siegmund, K.D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013, 41, e90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.-P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.T.; Hansen, K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [Green Version]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Wilhelm-Benartzi, C.S.; Koestler, D.C.; Karagas, M.R.; Flanagan, J.M.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J.; Houseman, E.A.; Brown, R. Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer 2013, 109, 1394–1402. [Google Scholar] [CrossRef] [Green Version]
- Jaffe, A.E.; Murakami, P.; Lee, H.; Leek, J.T.; Fallin, M.D.; Feinberg, A.P.; Irizarry, R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012, 41, 200–209. [Google Scholar] [CrossRef] [Green Version]
- Siegel, E.M.; Berglund, A.E.; Riggs, B.M.; Eschrich, S.A.; Putney, R.M.; Ajidahun, A.O.; Coppola, D.; Shibata, D. Expanding Epigenomics to Archived FFPE Tissues: An Evaluation of DNA Repair Methodologies. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2622–2631. [Google Scholar] [CrossRef] [Green Version]
- Siegel, E.M.; Eschrich, S.; Winter, K.; Riggs, B.; Berglund, A.; Ajidahun, A.; Simko, J.; Moughan, J.; Ajani, J.; Magliocco, A. Epigenomic Characterization of Locally Advanced Anal Cancer: A Radiation Therapy Oncology Group 98-11 Specimen Study. Dis. Colon Rectum 2014, 57, 941–957. [Google Scholar] [CrossRef]
- Konno, H.; Yamauchi, S.; Berglund, A.; Putney, R.M.; Mulé, J.J.; Barber, G.N. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 2018, 37, 2037–2051. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Hu, Z.; Mahal, B.A.; Zhao, S.D.; Kensler, K.H.; Pi, J.; Hu, X.; Zhang, Y.; Wang, Y.; Jiang, J.; et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell 2018, 34, 549–560.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Risk | High | Low | p-Value 1 |
|---|---|---|---|
| n = 11 | n = 13 | ||
| Age at diagnosis | 65.5 ± 4.1 | 59.4 ± 10.5 | 0.006 |
| PSA | 8.3 ± 6.59 | 9.23 ± 9.33 | 0.350 |
| Vital status | 0.500 | ||
| Deceased | 1 | 0 | |
| Alive | 10 | 11 | |
| Missing | 0 | 2 | |
| Gleason score | <0.0001 | ||
| 6 | 0 | 8 | |
| 7 (3 + 4) | 0 | 5 | |
| 7 (4 + 3) | 7 | 0 | |
| 8–9 | 4 | 0 | |
| Biochemical recurrence (BCR) | 0.160 | ||
| Yes | 3 | 1 | |
| No | 7 | 12 | |
| Missing | 1 | 0 | |
| Clinical Stage | 0.330 | ||
| T2a | 1 | 3 | |
| T2c | 6 | 9 | |
| T3a | 1 | 0 | |
| T3b | 3 | 1 | |
| Surgical margins | 0.480 | ||
| Yes | 1 | 0 | |
| No | 8 | 11 | |
| Missing | 2 | 2 | |
| Family history of prostate cancer | 0.830 | ||
| No | 3 | 5 | |
| Yes | 3 | 3 | |
| Missing | 5 | 5 |
| Probe ID | Gene Symbol | GeneBody | Chr | Position | p-Value | Delta β-Value | Mean (Low-Risk) | Mean (High-Risk) |
|---|---|---|---|---|---|---|---|---|
| cg22030684 | COL9A2 | Body | 1 | 40,781,708 | 0.000067 | −0.213457 | 0.449935 | 0.236478 |
| cg25161377 | FAM179A | Body | 2 | 29,237,783 | 0.000126 | −0.221753 | 0.654878 | 0.433124 |
| cg15218485 | RREB1 | Body | 6 | 7,201,665 | 0.000899 | 0.220195 | 0.238283 | 0.458478 |
| cg24163360 | SLC17A2 | TSS1500 | 6 | 25,931,557 | 0.000087 | −0.218759 | 0.685592 | 0.466833 |
| cg25641223 | PDE10A | Body | 6 | 165,747,945 | 0.000104 | −0.200032 | 0.640184 | 0.440152 |
| cg11747142 | FAM71F2 | TSS200 | 7 | 128,312,331 | 0.00054 | 0.229507 | 0.477201 | 0.706708 |
| cg17983571 | JMJD1C | Body | 10 | 65,186,953 | 0.000427 | 0.23785 | 0.470854 | 0.708704 |
| cg05363118 | PLEKHS1 | 5′UTR | 10 | 115,523,310 | 0.000081 | −0.206065 | 0.589143 | 0.383078 |
| cg05258834 | TNNI2 | Body | 11 | 1,862,477 | 0.000455 | −0.24573 | 0.648741 | 0.403012 |
| cg21359838 | OR51A4 | TSS1500 | 11 | 4,969,708 | 0.000186 | −0.212002 | 0.572201 | 0.360198 |
| cg25921194 | RNF169 | Body | 11 | 74,492,567 | 0.000161 | −0.24419 | 0.624767 | 0.380577 |
| cg19092163 | SPNS2 | Body | 17 | 4,403,417 | 0.00075 | −0.231457 | 0.433052 | 0.201596 |
| cg15002904 | ADAMTSL5 | Body | 19 | 1,510,692 | 0.000368 | −0.242087 | 0.496689 | 0.254602 |
| cg17713488 | COL5A3 | Body | 19 | 10,077,935 | 0.000157 | 0.360569 | 0.405099 | 0.765668 |
| cg22669123 | CYP4F12 | TSS1500 | 19 | 15,782,644 | 0.000677 | −0.215233 | 0.627928 | 0.412695 |
| cg09154639 | RAE1 | TSS1500 | 20 | 55,925,570 | 0.000905 | 0.226258 | 0.561288 | 0.787546 |
| cg14539730 | GABRQ | Body | X | 151,809,946 | 0.000227 | 0.389047 | 0.362807 | 0.751854 |
| Ancestral Population | Average | SD | Maximum | Minimum |
|---|---|---|---|---|
| African | 0.2408 | 0.2256 | 0.8576 | 0.0274 |
| European | 0.6420 | 0.2181 | 0.9716 | 0.0938 |
| Indigenous American | 0.1173 | 0.0897 | 0.2516 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Deya, G.; Matta, J.; Encarnación-Medina, J.; Ortiz-Sanchéz, C.; Dutil, J.; Putney, R.; Berglund, A.; Dhillon, J.; Kim, Y.; Park, J.Y. Differential DNA Methylation in Prostate Tumors from Puerto Rican Men. Int. J. Mol. Sci. 2021, 22, 733. https://doi.org/10.3390/ijms22020733
Ruiz-Deya G, Matta J, Encarnación-Medina J, Ortiz-Sanchéz C, Dutil J, Putney R, Berglund A, Dhillon J, Kim Y, Park JY. Differential DNA Methylation in Prostate Tumors from Puerto Rican Men. International Journal of Molecular Sciences. 2021; 22(2):733. https://doi.org/10.3390/ijms22020733
Chicago/Turabian StyleRuiz-Deya, Gilberto, Jaime Matta, Jarline Encarnación-Medina, Carmen Ortiz-Sanchéz, Julie Dutil, Ryan Putney, Anders Berglund, Jasreman Dhillon, Youngchul Kim, and Jong Y. Park. 2021. "Differential DNA Methylation in Prostate Tumors from Puerto Rican Men" International Journal of Molecular Sciences 22, no. 2: 733. https://doi.org/10.3390/ijms22020733
APA StyleRuiz-Deya, G., Matta, J., Encarnación-Medina, J., Ortiz-Sanchéz, C., Dutil, J., Putney, R., Berglund, A., Dhillon, J., Kim, Y., & Park, J. Y. (2021). Differential DNA Methylation in Prostate Tumors from Puerto Rican Men. International Journal of Molecular Sciences, 22(2), 733. https://doi.org/10.3390/ijms22020733






