Accumulation of Mitochondrial RPPH1 RNA Is Associated with Cellular Senescence
Abstract
1. Introduction
2. Results
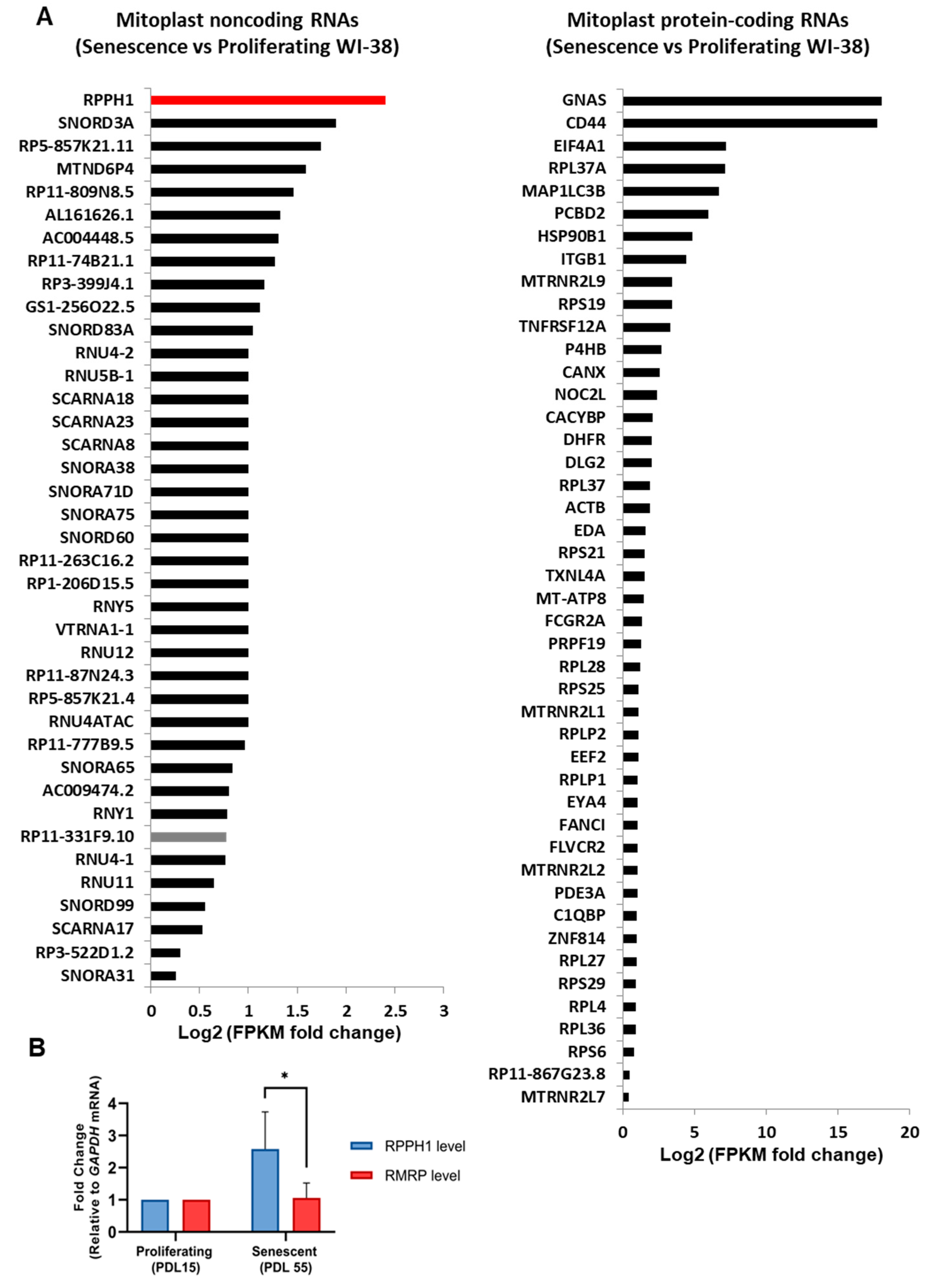
2.1. Expression Changes of Mitochondrial RNAs in Cellular Senescence
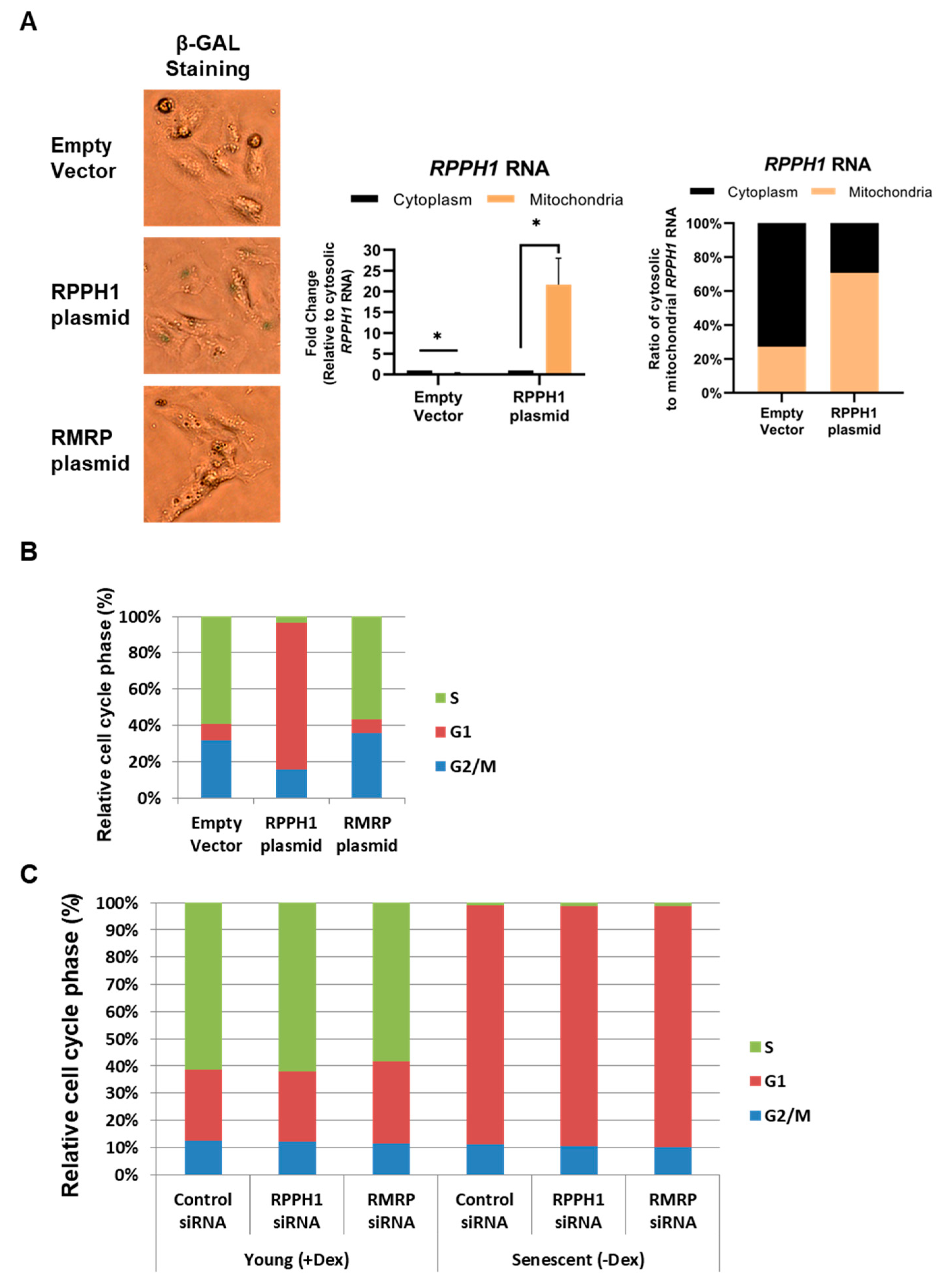
2.2. A Senescence-Associated Mitochondrial Localization of RPPH1 RNA Is Modulated by AUF1
2.3. Replicative Cellular Senescence Is in Part Attributed to Mitochondrial RPPH1 Overexpression
3. Discussion
3.1. Senescence-Associated Mitochondrial Noncoding RNAs
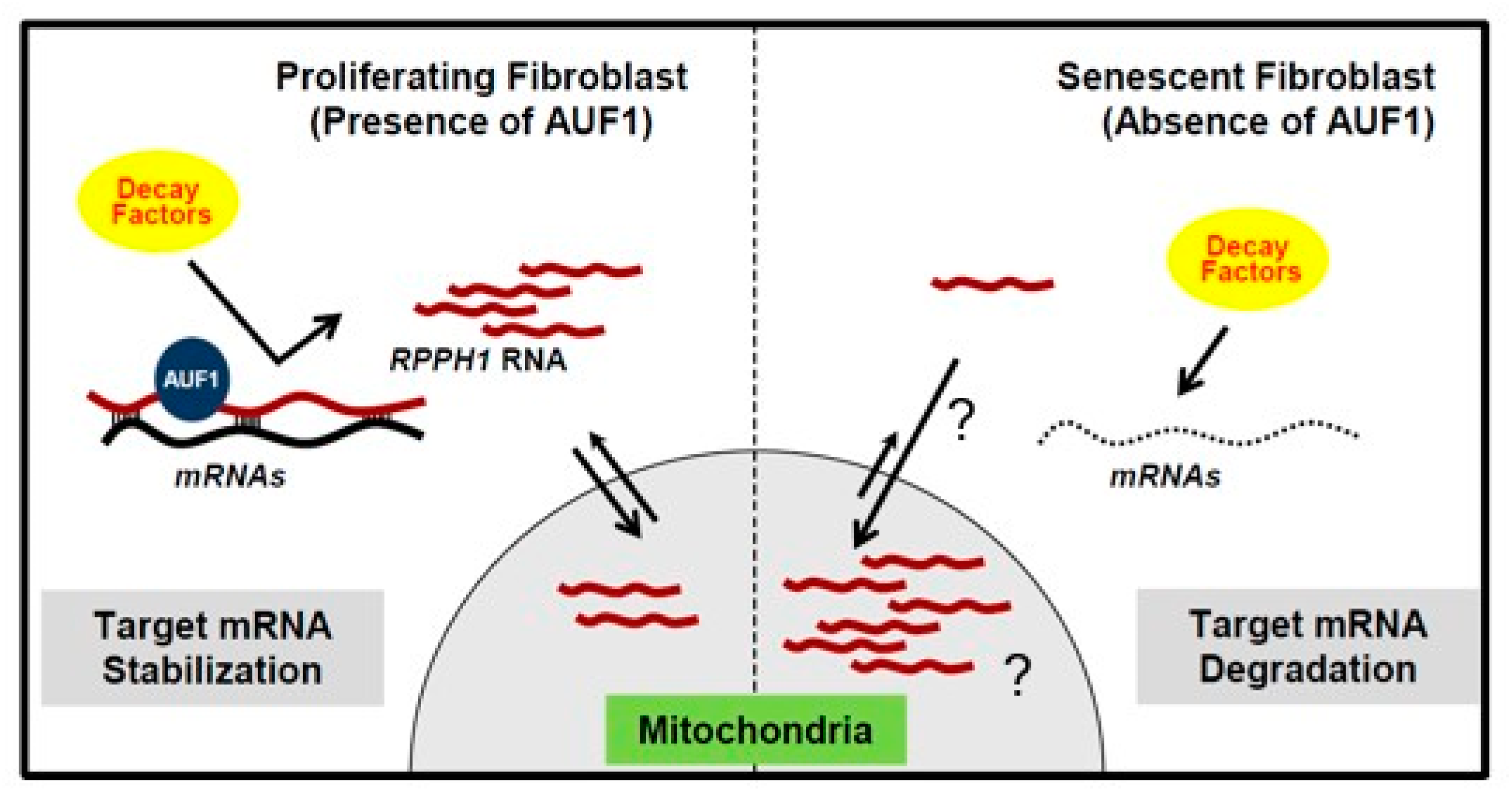
3.2. Senescence-Induced RPPH1 RNA Accumulation in Mitochondria
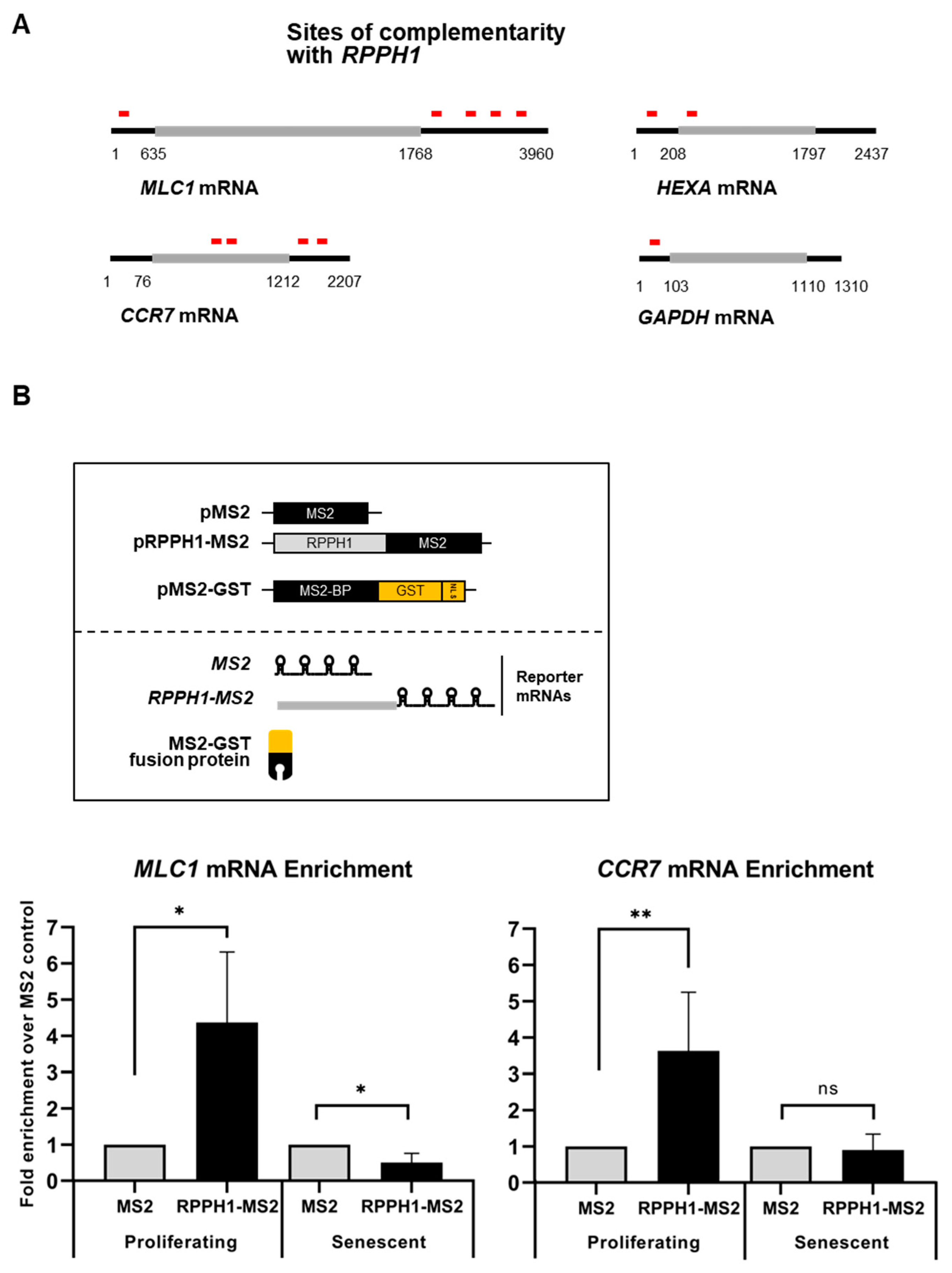
3.3. Target mRNA Stabilization by Partial RNA Complementarity
4. Material and Methods
4.1. Cell Culture, Transfection, Small Interfering RNAs, and Plasmids
4.2. Western Blot Analysis
4.3. RNP Analysis
4.4. RNA Quantification by qPCR
4.5. Subcellular Fractionation
4.6. Bioinformatic Analysis of RPPH1 RNA Interaction Sites with mRNAs
4.7. Cellular Senescence Assay
4.8. Luciferase Reporter Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voss, T.C.; Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014, 15, 69–81. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed]
- van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Zarnack, K.; Luscombe, N.M.; Ule, J. Protein–RNA interactions: New genomic technologies and perspectives. Nat. Rev. Genet. 2012, 13, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Izaurralde, E. Breakers and blockers—miRNAs at work. Science 2015, 349, 380–382. [Google Scholar] [CrossRef] [PubMed]
- Zealy, R.W.; Fomin, M.; Davila, S.; Makowsky, D.; Thigpen, H.; McDowell, C.H.; Cummings, J.C.; Lee, E.S.; Kwon, S.H.; Min, K.W.; et al. Long noncoding RNA complementarity and target transcripts abundance. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Tollervey, D. Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nat. Protoc. 2014, 9, 711–728. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.H. The utility of protein and mRNA correlation. Trends Biochem. Sci. 2015, 40, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Parker, R. RNA degradation in Saccharomyces cerevisae. Genetics 2012, 191, 671–702. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Lorsch, J.R. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, a011544. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.; Parker, R. Quality control: Is there quality control of localized mRNAs? J. Cell Biol. 2014, 204, 863–868. [Google Scholar] [CrossRef][Green Version]
- Martin, K.C.; Ephrussi, A. mRNA localization: Gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef]
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef]
- Buchan, J.R. mRNP granules. RNA Biol. 2014, 11, 1019–1030. [Google Scholar] [CrossRef]
- Weis, B.L.; Schleiff, E.; Zerges, W. Protein targeting to subcellular organelles via mRNA localization. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 260–273. [Google Scholar] [CrossRef]
- Mili, S.; Moissoglu, K.; Macara, I.G. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 2008, 453, 115–119. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Nicchitta, C.V. Primary Role for Endoplasmic Reticulum-bound Ribosomes in Cellular Translation Identified by Ribosome Profiling. J. Biol. Chem. 2012, 287, 5518–5527. [Google Scholar] [CrossRef]
- Reid, D.W.; Chen, Q.; Tay, A.S.L.; Shenolikar, S.; Nicchitta, C.V. The Unfolded Protein Response Triggers Selective mRNA Release from the Endoplasmic Reticulum. Cell 2014, 158, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W.; Potuschak, T. Difference between Mitochondrial RNase P and Nuclear RNase P. Mol. Cell. Biol. 2001, 21, 8236–8237. [Google Scholar] [CrossRef]
- Bartkiewicz, M.; Gold, H.; Altman, S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989, 3, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Srikantan, S.; Gorospe, M. MS2-TRAP (MS2-tagged RNA affinity purification): Tagging RNA to identify associated miRNAs. Methods 2012, 58, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.; Kang, M.J.; Xu, J.; Selimyan, R.; Yoon, J.H.; Martindale, J.L.; De, S.; Wood, W.H., 3rd; Becker, K.G.; et al. Senescence-associated lncRNAs: Senescence-associated long noncoding RNAs. Aging Cell 2013, 12, 890–900. [Google Scholar] [CrossRef]
- Yoon, J.H.; De, S.; Srikantan, S.; Abdelmohsen, K.; Grammatikakis, I.; Kim, J.; Kim, K.M.; Noh, J.H.; White, E.J.; Martindale, J.L.; et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014, 5, 5248. [Google Scholar] [CrossRef]
- Holzmann, J.; Frank, P.; Löffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and Functional Reconstitution of the Human Mitochondrial tRNA Processing Enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019, 47, 7294–7305. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; Richter, J.D. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008, 22, 3449–3460. [Google Scholar] [CrossRef]
- Srikantan, S.; Gorospe, M.; Abdelmohsen, K. Senescence-associated microRNAs linked to tumorigenesis. Cell Cycle 2011, 10, 3211–3212. [Google Scholar] [CrossRef][Green Version]
- Laberge, R.-M.; Sun, Y.; Orjalo, A.V.; Patil, C.K.; Freund, A.; Zhou, L.; Curran, S.C.; Davalos, A.R.; Wilson-Edell, K.A.; Liu, S.; et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 2015, 17, 1049–1061. [Google Scholar] [CrossRef]
- Herranz, N.; Gallage, S.; Mellone, M.; Wuestefeld, T.; Klotz, S.; Hanley, C.J.; Raguz, S.; Acosta, J.C.; Innes, A.J.; Banito, A.; et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 2015, 17, 1205–1217. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St. Laurent Iii, G.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Niu, S.; Tan, M.; Huang, C.; Li, M.; Song, Y.; Wang, Q.; Chen, J.; Shi, S.; Lan, P.; et al. Cryo-EM Structure of the Human Ribonuclease P Holoenzyme. Cell 2018, 175, 1393–1404.e11. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.W.; Chun, Y.L.; Kim, A.Y.; Lloyd, L.T.; Ko, S.; Yoon, J.-H.; Min, K.-W. Accumulation of Mitochondrial RPPH1 RNA Is Associated with Cellular Senescence. Int. J. Mol. Sci. 2021, 22, 782. https://doi.org/10.3390/ijms22020782
Lee JW, Chun YL, Kim AY, Lloyd LT, Ko S, Yoon J-H, Min K-W. Accumulation of Mitochondrial RPPH1 RNA Is Associated with Cellular Senescence. International Journal of Molecular Sciences. 2021; 22(2):782. https://doi.org/10.3390/ijms22020782
Chicago/Turabian StyleLee, Ji Won, Yoo Lim Chun, Ah Young Kim, Lawson T. Lloyd, Seungbeom Ko, Je-Hyun Yoon, and Kyung-Won Min. 2021. "Accumulation of Mitochondrial RPPH1 RNA Is Associated with Cellular Senescence" International Journal of Molecular Sciences 22, no. 2: 782. https://doi.org/10.3390/ijms22020782
APA StyleLee, J. W., Chun, Y. L., Kim, A. Y., Lloyd, L. T., Ko, S., Yoon, J.-H., & Min, K.-W. (2021). Accumulation of Mitochondrial RPPH1 RNA Is Associated with Cellular Senescence. International Journal of Molecular Sciences, 22(2), 782. https://doi.org/10.3390/ijms22020782






