1. Introduction
The molecular processes preceding and optimizing the activation of store-operated calcium entry (SOCE) are difficult to study because they require single molecule resolution, while at the same time, the interaction of a multiple of proteins needs to be examined [
1,
2]. After several early reports supported tetrameric ORAI1 conformations [
3,
4,
5,
6], the current view is that ORAI1 proteins assemble as a hexameric channel complex at rest [
7,
8,
9] and that these hexameric protein complexes are randomly distributed throughout the plasma membrane. Upon Ca
2+ store depletion, STIM proteins redistribute in the endoplasmic reticulum (ER) membrane towards the cytosolic side of ER plasma membrane contact zones, where typical dense STIM1 accumulations known as puncta are formed [
10,
11]. In these junctional areas between the ER and plasma membrane, STIM1 proteins approach the plasma membrane on the cytoplasmic side where they interact with ORAI1, resulting in a conformational change that opens the ORAI1 channels. Yet, how the ORAI1 channels translocate to these regions in order to get trapped there is not fully understood. It is assumed that single ORAI1 channels randomly diffuse within the plasma membrane until they arrive at sites of activated STIM1, followed by the binding and activation of ORAI1 Ca
2+ channels, leading to SOCE [
2]. Different mechanisms may play a role, for example, the additional insertion of ORAI1 from intracellular stores into the regions of puncta [
12], or potentially a pre-clustering of several hexamers which are then pulled in or trapped at the ER-PM junctions.
By using liquid phase electron microscopy (LPEM) capable of studying ORAI1 proteins in the plasma membrane of intact cells [
9,
13], we set out to examine the difference in spatial distribution of ORAI1 between cells at rest and upon SOCE-activation. Our aim was to test: (1) if single ORAI1 channels are indeed randomly distributed at rest, (2) to characterize, with nanoscale resolution, how the distribution of ORAI1 differs in activated cells, including their redistribution into puncta. For this purpose, ORAI1 proteins were expressed with a stoichiometric expression of cytosolic green fluorescent protein (GFP) and with a hemagglutinin (HA) tag located in the extracellular loop between transmembrane regions (TM) 3 and 4 in HEK cells and labeled with quantum dots (QDs). A QD provides both a fluorescence label for light microscopy and a nanoparticle label for detection with electron microscopy [
9,
13].
Figure 1A depicts the dimensions of an ORAI1 hexamer [
7] in a hypothetical arrangement of two QDs labels bound to the same ORAI1 hexamer (see also scheme in
Figure S1, showing an alternative view of an ORAI1-label configuration). Due to the flexibility of the linker, the center-to-center distance between both QDs may vary between 10 and 35 nm. Spatial patterns of individual QD labels on whole cells were imaged under resting conditions and after two levels of SOCE-activation. The spatial distributions of the labeled ORAI1 proteins were subsequently analyzed with nanometer spatial resolution, as obtained with the LPEM method [
9,
14].
3. Discussion
Our study aimed to elucidate how the spatial organization of ORAI1 proteins in the plasma membrane changes upon Ca
2+ channel activation. We have used an HA-tagged ORAI1 protein in combination with a two-step QD labeling approach, resulting in a label-target ratio of <1, thereby excluding any clustering of more than one QD at a single ORAI1 protein [
9]. It can also be excluded that the applied QD-labeling protocol induces artificial clustering of membrane proteins. This proof was published for a similar QD-labeling approach of another membrane protein, namely HER2, where randomly distributed QD label positions occurred in a phenotypic distinct subpopulation of HER2 overexpressing cancer cells [
23,
24]. The proteins were detected in the intact and hydrated cell, using correlative light microscopy and LPEM, yielding the position information of several hundred thousand single-labeled ORAI1 proteins.
The fact that we analyzed only a fraction of ORAI1 proteins, namely those with a bound QD, leaving the majority of all ORAI1 unlabeled, and thus “invisible”, is not hampering or biasing our analysis of the spatial distribution of ORAI1, because we can reasonably assume the labeling process to be stochastic. This randomness allows us to consider the labeled fraction of ORAI1 as representative of all present ORAI1, independently of the relative size of this fraction and the labeling efficiency. Fractional random labeling and microscopic analysis of a representative share of the target proteins are widely used in cell biology to study the spatial localization of single proteins. Mostly, the labeling efficiency, or the share of analyzed fluorescent labels, is even significantly lower than our estimated labeling efficiency [
25].
Studying engineered proteins via cell expression systems may raise concerns about potential mismatches between the studied situation and that in native tissue cells, as the protein under investigation may be expressed at a too high level compared to the other endogenous proteins interacting. An imbalance between the ratios of the interacting protein species could in principle also influence the spatial distribution of ORAI1 in the plasma membrane. To evaluate if we had unphysiologically high levels of ORAI1 in our cellular expression system, we calculated the average surface density of the truly present ORAI1 channels. Supposing hexameric channels and applying an estimated labeling efficiency of 30% resulted in an average ORAI1 channel surface density of ~75/μm
2, the surface density decreased to ~50/μm
2 in the separately analyzed group of low expressing cells, whereas in the high expressing cells it reached ~120/μm
2. Comparing this range of values with the published surface density of presumably endogenous ORAI1 channels in regular HEK cells, amounting to 193 ± 91/μm
2 [
26], indicates that our ORAI1 expression levels were in a lower physiological range, at least for HEK cells. Note that endogenous ORAI1 and ORAI2 were almost abolished in the used modified HEK cell line. Nevertheless, one has to be cautious to generally extrapolate our results to cells from other tissues, because large variations in relative expression ratios of ORAI1 and its interacting proteins exist between different tissues and cell types.
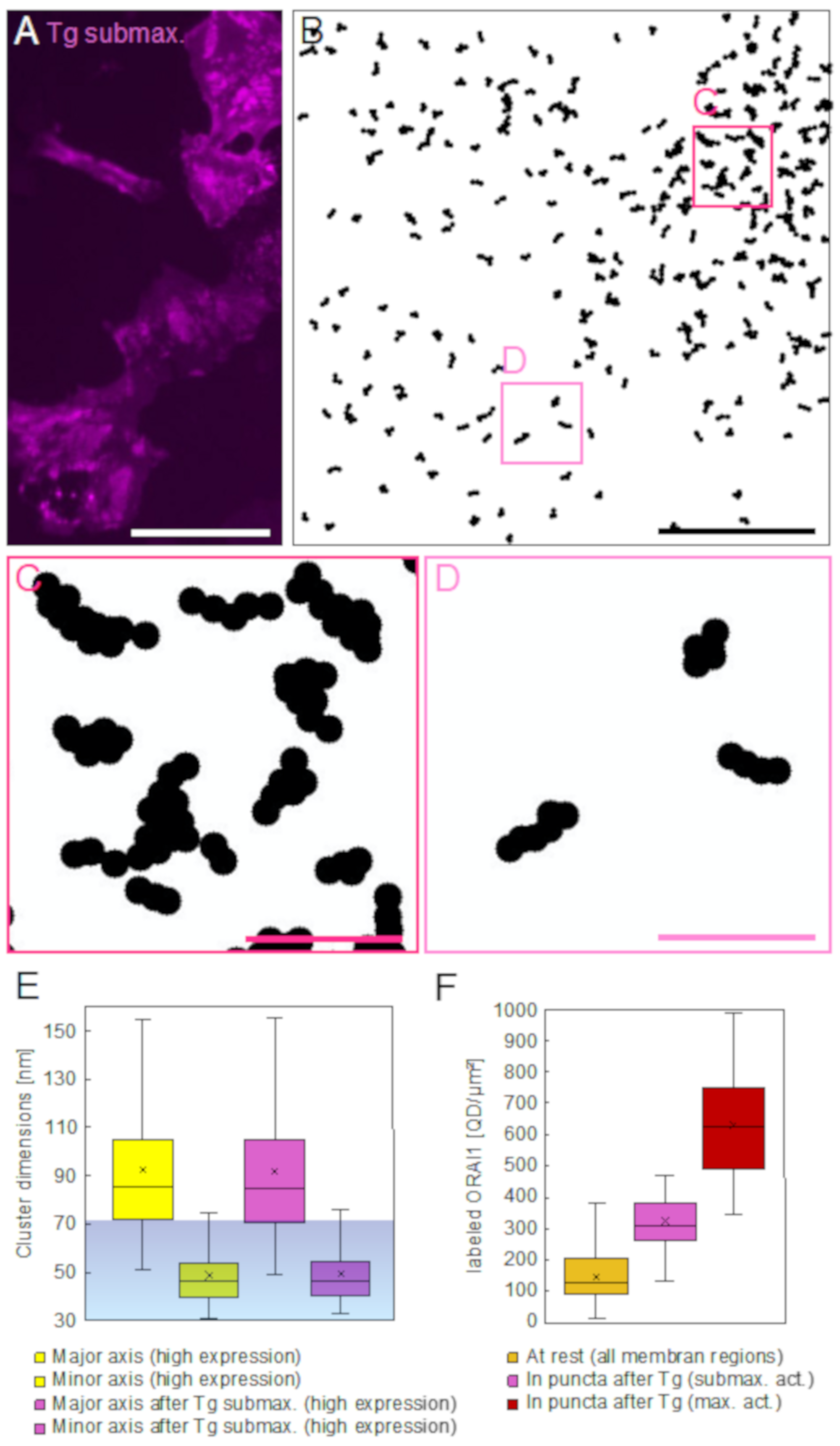
In resting cells, we discovered that ORAI1 often assembled in chain-like structures extending up to 130 nm. ORAI1 proteins thus tended to form supra-molecular arrangements. Considering a predominant distance of ~15 nm between adjacent ORAI1 channels in puncta, as measured by electron microscopy [
26], the underlying chain-like conformation could contain between two and eight ORAI1 channels. The existence of such multi-channel ORAI1 conformations contradicts the assumption that ORAI1 channels predominantly exist as independent entities, randomly distributed in the plasma membrane [
18,
26]; those authors further suggested that single channels were freely diffusing within these patches. Although they also used electron microscopy to examine overexpressed ORAI1, the proteins were imaged without labels, and in freeze-fractured cells [
26], posing challenges for the discrimination of ORAI against other neighboring membrane proteins, while labeling as used by us ensures specific detection of ORAI and thus provides more reliable information about ORAI’s spatial distribution. Nevertheless, those authors stated the occasional detection of ORAI1 channels in specific arrangements of pairs and short chains.
In the STEM images recorded after SOCE-activation, ORAI1 clusters had been trapped into the regions of puncta where, depending on the availability of STIM proteins, they often aligned side-by-side. This was found after sub-maximal activation, or condensed further until reaching a rather confluent pattern, after maximal activation. Measurements of ORAI1 densities in maximally activated cells, inside and outside puncta, revealed on average a four-fold increase within the regions of puncta achieved by the redistribution of a minor ORAI1 protein fraction, ranging between 14% and 24% of all plasma membrane-bound ORAI1. Our results do not support earlier findings reporting the insertion of ORAI1 from intracellular pools into puncta after SOCE-activation [
12]. Such a replenishment of plasma membrane localized ORAI1 would have led to an increase in the average ORAI1 surface density in SOCE-activated cells versus cells at rest; however, the average ORAI1 densities in our experiments remained similar. A possible reason for this discrepancy might be the exclusive use of formaldehyde as a fixation agent in this earlier study, which is known to insufficiently fix membrane proteins [
27,
28] and to induce artificial redistributions of ORAI1 proteins [
29].
In resting cells, Perni et al. reported micron-sized patches with ragged edges, assuming these areas contained dense accumulations of ORAI1 channels [
26]. We could not confirm this finding; instead, labeled ORAI1 distributed rather evenly throughout the plasma membrane of resting cells with mono-dispersed and clustered label formations. This discrepancy might lie in differences between the preparation and imaging techniques used, notably the lack of identifying labels, as already mentioned above. Our comparison between the cluster analysis of STEM image data with their density-matching simulations led us to propose that a significant fraction of the ORAI channels formed linear arrangements. For all examined experimental groups, the dimensions and aspect ratios of labeled-ORAI1 clusters >2 in the STEM images were significantly larger (
p < 0.001) than those of clusters >2 found in their matching simulations (see
Figures S5 and S6). High-order aggregates of non-solubilized ORAI proteins were found earlier with the biochemical technique of Native Gel Electrophoresis using perfluorooctanoic acid (PFO), but these results were interpreted as technical artifacts [
5]. Furthermore, ORAI1 diffusion coefficients were reported to exhibit a large range of different values, whereby most ORAI1 proteins had sub-diffusive velocities [
2] possibly caused by differently sized molecular ORAI1 assemblies [
30], a finding which fits the idea of supra-molecular ORAI1 channel arrangements.
Concerning the larger, peculiar 2D assembly patterns detected in SOCE-activated cells, such as micrometer-long strands and ring-like structures, we were unable to find matching reports in the literature. Perni et al. did confirm the presence of dense ORAI accumulations in puncta after SOCE-activation, but their observations did not include strand- and ring-like 2D structures. This discrepancy is likely due to the small width of these strands, often of a single labeled ORAI1, and the associated difficulty of discerning such small features amongst the other neighboring proteins in the crowded environment of the plasma membrane without using labels.
We propose that a possible function of clustering of ORAI1 in supramolecular arrangements could be a more efficient concentration process of ORAI1 Ca
2+ channels in puncta upon SOCE activation compared to a mechanism via the collection of single channels. The capturing and dragging of only one ORAI1 protein belonging to a supramolecular cluster will drag all the others with it until the whole cluster reaches the ER plasma membrane contact zone. A confluent filling of the plasma membrane areas located above accumulated STIM in the ER plasma membrane contact zones near supra-molecular ORAI1 clusters, rather than single ORAI1 channels, would not only be faster but would also help in achieving a more homogenous filling. Additionally, this would prevent jamming of ORAI1 channels at the periphery of the puncta, which would hinder and delay filling of the more central areas. An amplifying effect, due to the import of supra-molecular ORAI clusters instead of single ORAI1 channels, would lower the required amount of effectively trapped and transported ORAI1 far below the 14% to 24% of ORAI1 proteins found to be redistributed after maximal SOCE-activation. Such an amplifying effect would, for instance, play an important role in the immune system by accelerating the formation of immune synapses between T cells and antigen-presenting cells [
31]. The comparison of ORAI1 distributions in accumulation zones in sub-maximally activated cells revealed less dense accumulations of ORAI1 clusters than in maximally activated cells. Yet, the accumulation areas showed many aligned supra-molecular ORAI1 clusters, further supporting the concept that the import of preformed, supra-molecular ORAI1 clusters contributed to the accumulation of ORAI1 in puncta. The existence of such an amplifying mechanism could be tested in the future, for example, using dynamic nanoscale microscopy provided by advanced super resolution fluorescence microscopy or by LPEM enhanced for time-resolved imaging at ultra-low electron dose, which is albeit not yet available [
32].
The existence of supra-molecular protein clusters in the plasma membrane, with similar dimensions and numbers of involved proteins as detected in this study for ORAI1 proteins, possibly points to a general organization principle in the plasma membrane, as such protein arrangements were also found for other membrane proteins, mainly by using super-resolution fluorescence microscopy. After over-expression in HEK cells, for instance, about half of the N-methyl-D-aspartate receptor receptors were found in clusters containing up to 12 receptors [
33]. Also, for the family of G-protein coupled receptors, about half of the μ-opioid receptors, and 85% of κ-opioid receptors were reported to reside in clusters with dimensions of 80 nm to 100 nm, comprising on average eight and nine receptors, respectively [
34]. Assemblies of 10 or more receptors were also reported for the epidermal growth factor receptor (EGFR) [
35], and chains containing six or more receptors for the closely related human epidermal growth factor receptor 2 (HER2) [
36], which represents an example in which endogenous proteins were labeled.
It is not known if a linking protein responsible for the organization of supra-molecular ORAI1 clusters exists. STIM proteins fulfill such an ORAI1 binding function and concomitant to binding activate ORAI1 channels in the puncta. Though outside these regions and at rest, clustering of ORAI1 due to STIM can be excluded, as shown by control experiments (see
Figure S2). The possible mechanism behind the clustering of ORAI1 could involve homo-oligomerization [
37,
38], the involvement of cytoskeletal proteins [
39,
40], or septins, recently shown to be involved in cellular calcium signaling through ORAI1 [
30], as well as a range of other known ORAI1-binding proteins [
41,
42].
4. Materials and Methods
Fetal bovine serum (FBS), 2-mercaptoethanol (ME), Tg, cyclopiazonic acid, and sodium azide, were either from Fisher Scientific, Schwerte, Germany, or Sigma Aldrich, Darmstadt, Germany. ScreenFect®A Transfection reagent was from Incella GmbH, Eggenstein-Leopoldshafen, Germany. Anti-HA-biotin, high affinity (3F10) from rat IgG1 was from Roche Diagnostics, Mannheim, Germany. Dulbecco’s phosphate buffered saline (DPBS), Modified Eagle’s Medium (MEM), normal goat serum (GS), CellStripper and QD Qdot® 655, and Qdot®565 streptavidin conjugates were from Fisher Scientific GmbH, Schwerte, Germany. ROTISOLV® high pressure liquid chromatography grade pure water, acetone and ethanol, phosphate buffered saline (PBS) 10 × solution, electron microscopy grade glutaraldehyde (GA) 25% solution, D-saccharose, sodium chloride, glycine, biotin free and molecular biology grade albumin fraction V (BSA), and sodium cacodylate trihydrate were from Carl Roth GmbH + Co. KG, Karlsruhe, Germany. Electron microscopy grade formaldehyde (FA) 16% solution was from Science Services GmbH, Munich, Germany. The 0.01% poly-L-Lysine (PLL) solution (mol wt. 70,000–150,000), sodium tetraborate, and boric acid were from Sigma-Aldrich Chemie GmbH, Munich, Germany. CELLVIEW cell culture dishes (35 mm) with four compartments and glass bottoms were from Greiner Bio-One GmbH, Frickenhausen, Germany. Custom designed silicon microchips were purchased from DENSsolutions, Delft, Netherlands. The microchips had outer dimensions of 2.0 × 2.6 × 0.4 mm and each contained a silicon nitride (SiN) membrane window, usually with dimensions of 150 × 400 µm (sometimes larger) along with a membrane of 50 nm thickness. Trivial transfer multilayer graphene was purchased from ACS Material LLC, Pasadena, CA, USA. NaCl2 crystals were from Plano GmbH, Wetzlar, Germany.
HEK293 cells were obtained from ATCC, Wesel, Germany, and genetically modified using CRISPR/Cas9-mediated gene deletion of endogenous ORAI1-2 genes (named CRI_1), or ORAI1-3 genes (named CRI_2, which were used for controls shown in the
Supplementary information), as described in [
9]. A HEK cell line lacking endogenous STIM proteins (CRI_STIM) [
19] was obtained from Donald L. Gill [
19]. An ORAI1 construct with an extracellular nine amino acid HA-tag within the second extracellular loop of ORAI1 was used for labeling the protein with a QD [
43]. The cDNA sequence encoding for HA tag is TACCCATATGACGTACCGGATTACGCC which translates to the amino acid sequence YPYDVPDYA. The DNA construct also contained a DNA encoding green fluorescent protein (GFP) but separated by a cleavable peptide sequence (P2A) thus guaranteeing the same expression ratio [
13,
44]. The ORAI1-GFP DNA construct was transiently expressed in the HEK cells using ScreenFect
®A as described in [
9], which also includes the results of functional tests of the transfected ORAI1-HA DNA constructs. All constructs encode human sequences.
4.1. Preparation of Microchips with Transfected HEK Cells
CELLVIEW dishes and microchips with thin SiN windows were used as a support for CRI_1 cells. Preparation of new microchips was performed as described previously [
45], briefly, protective photoresist coating on the microchips was removed with acetone and ethanol, the microchips and dishes were plasma-cleaned for 5 min, coated with PLL, and immersed/filled with cell medium (supplemented with 10% FBS and 50 μM ME). Cells grown in a 25 cm
2 flask were harvested at ~90% confluency and washed once in supplemented cell medium. Cell transfection was performed according to the supplier’s instructions. In experiments with ORAI1 at rest, 0.25 μg ORAI1-HA DNA and 1.5 μL transfection reagent (TR) were used per compartment of a four-compartment dish (35 mm diameter) containing a final volume of 420 μL cell suspension. In experiments with activated ORAI1, 0.17 μg ORAI1-HA DNA, 0.55 μg STIM1-mcherry DNA to yield a 1:3 ratio between ORAI1 and STIM1, or 0.17 μg for each DNA and 2 μL TR for a 1:1 ratio, were used per dish compartment. The respective volumes for single microchips kept in wells of a 96-well plate were 25% of those used for dish compartments. The cell samples were then incubated for 24 h in the CO
2 incubator.
4.2. Labeling of Overexpressed ORAI1-HA at Rest or after Activation
A two-step labeling protocol [
13,
45] was applied using a biotinylated anti-HA Fab, followed by labeling with 20 nM streptavidin-conjugated QD, as previously described [
9]. After transfection, the samples were rinsed with supplemented cell medium pre-warmed to 37 °C. For experiments examining unstimulated ORAI1 distribution, in the following named cells at rest, the samples were briefly rinsed in pre-warmed 0.1 M cacodyl buffer (CB) containing 0.1 M sucrose, pH 7.4, and incubated in 3% formaldehyde/0.2% glutaraldehyde in CB for 10 min at room temperature, thereby assuring fast and permanent fixation of membrane proteins, eliminating their diffusion [
27,
46,
47]. For the examination of activated ORAI, cells were first rinsed twice with pre-warmed medium (MEM, suppl. with 10% FCS and 50 μM ME), then incubated with 1 μM Tg, or 30 μM CPA (both in supplemented medium), for 15 min at 37 °C (in the CO
2 incubator). The activated cells were rinsed once with pre-warmed medium, and once with pre-warmed CB, followed by fixation as described above. Fixation was terminated by rinsing once with CB, three times with PBS, 2 min of incubation in GLY-PBS (0.1% glycine in PBS) for 2 min, followed by a rinse in PBS. The cells were then incubated in 400 ng/mL Anti-HA-Fab-Biotin labeling solution in PBS, first for 1 h at room temperature, followed by 3–6 h at 4 °C. The QD-labeling solutions were prepared by first diluting 1 µM Streptavidin-QD stock solutions 1:5 in 40 mM Borate buffer, pH 8.3, and a further dilution in hBSA-PBS (PBS with 1% BSA) to obtain a 20 nM QD labeling solution. After three times rinsing in PBS, cells were incubated in the streptavidin-QD labeling solutions for 12 min at room temperature. As the presence of BSA in the labeling solutions led to non-specific binding, it was omitted in the processing steps before the QD-incubation. After the QD incubation, the cells were rinsed four times with hBSA-PBS before fluorescence microscopy was performed.
4.3. Fluorescence Microscopy
After QD incubation and before the second fixation step, cells on microchips were imaged with an inverted fluorescence microscope (DMI6000B Leica, Germany) in a pristine, 35 mm cell culture glass-bottom dish filled with 2 mL hBSA-PBS. 20 × and 40 × objectives were used together with four channels, one collecting direct interference contrast (DIC), and three fluorescence channels for, GFP (460–520 nm excitation, 515–560 nm emission), QD655 (340–380 nm excitation, >420 nm emission), and QD565 (540–560 nm excitation, 580–620 emission).
4.4. Processing of Samples for LPEM
To stabilize the cells on the microchips for electron microscopy, the previously labeled samples were further fixed with 2% glutaraldehyde in CB for 10 min at room temperature. After one rinse with CB and three rinses with hBSA-PBS, they were stored in hBSA-PBS supplemented with 0.02% sodium azide at 4 °C until liquid-phase STEM, usually performed within 1–3 weeks. To keep the ORAI channels in their almost native environment, as provided by the imaging of hydrated intact cells, the microchip samples were covered with graphene. Multi-layer (3 to 5 layers) graphene on polymer was cleaned and transferred onto the sample as described previously [
14,
48]. For the coating of a microchip sample, a graphene sheet of approximately the size of a microchip was detached from its supporting NaCl crystal through immersion in a beaker filled with HPLC-grade water, placed under a binocular. The (wet) microchip was grabbed with a pair of fine-tipped tweezers, rinsed twice with pure water, and immersed in the liquid below the floating graphene. The graphene was then carefully scooped up by slowly drawing the microchip up and out of the water. The tweezers tips, still holding the microchip with the on top swimming graphene sheet, were fixed with a small rubber O-ring, and the other tweezer end was clamped into a small stand so that the microchip would hang free in the air. After a few minutes of drying time, the water on the graphene had evaporated and the graphene was directly adhering to the underlying still hydrated cells. A detailed description of this step can be found elsewhere [
14].
4.5. LPEM of QD-Labeled Whole Cells
To observe the individual QD-labeled ORAI1-HA positions, the graphene-coated samples were imaged in a transmission electron microscope (ARM 200, JEOL, Japan), with STEM dark field mode [
14]. The following settings were used: electron energy of 200 kV and 175 pA probe current. For orientation purposes, the imaging session started with the recording of two to three overview STEM images, covering the entire SiN-window area of the microchip with cells. These low magnification images were directly compared to the previously recorded fluorescence and DIC images and served to navigate to the selected representative cells chosen for high-magnification imaging. Several images from selected QD-labeled cells were recorded from randomly chosen plasma membrane regions. Note that central areas in the thickest nucleus containing part of the cells were excluded. Images of QD655 labeled cells were recorded with magnifications of mostly 60.000 ×, while 120.000 × was used for QD565 labeled cells (occasionally, other magnifications ranging between 40,000 and 150,000 × were applied), resulting in a pixel size of 1.6 nm and 0.8 nm for QD655 images and QD565 images, respectively. The image size of the 16-bit images was 2048 × 2048 pixels, comprising a scanning area of 10.1 µm
2 per 60.000 × magnified image, or 2.9 µm
2 per 120.000 × magnified image. The used pixel dwell-times ranged between 8 and 14 µs. The calculated electron doses for images of cells labeled with QD655 was usually between 40 and 60 e
−/Å
2, cells labeled with the smaller QD565 (shown in the
supplementary information) had maximally a dose of 125 e
−/Å
2, which is below the reported limit of radiation damage for such samples [
49].
4.6. Particle Detection
In order to obtain the lateral coordinates of the QD labels, all STEM images were first visually screened for infrequently occurring contaminants on the graphene (remnants of the production process) with dimensions and contrast characteristics sometimes hampering the automated label detection. In such cases, ImageJ (version 1.52a, NIH) was used to manually blank the respective contaminant in the image by covering them with a fitting shape, filled with the grey value of the surrounding background. QD labels were detected and localized by applying a dedicated plugin of local design in ImageJ described elsewhere [
23]. The main processing steps consisted of a Fourier filter for spatial frequencies between a factor of three smaller and a factor of three higher than the set size (7 nm), and a binarization with an automated threshold with a maximum entropy setting. The particles were automatically detected using the “Find Particles” tool, with a precision corresponding to the pixel size of 1.6 nm. A demonstration of the particle detection technique can be found in an earlier publication, including also an error estimation [
16].
For a better visual perception of the arrangement of labeled ORAI in selected images, we implemented another plugin of local design for the purpose of improved visual pattern recognition of the arranged labels. This was achieved by using the x-y center position data from the detected QD labels in a STEM image, and drawing black circles around each center position, on a blank image background of the same dimensions as the STEM image. Each circle thereby indicated the plasma membrane area where the underlying ORAI1 protein and bound labels localized. The size of the circle was determined by the dimensions of the QDs and the flexible linker, including the conjugated streptavidin, and the Anti-HA Fab. As can be seen in the drawn-to-scale model shown in
Figure 1 and
Figure S1, the maximal distance between the center of a QD and the ORAI1 protein is ~17 nm, so that the diameter of the circle was set to 35 nm.
4.7. Analysis of Supra-Molecular ORAI1 Arrangements and Punctae
For the detection of possible ORAI1 supramolecular arrangements, the processed STEM images, showing the labels as 35 nm diameter black circles, were analyzed with the “Analyze Particles” tool of ImageJ. The size limit was set such that clusters consisting of more than two labels were detected. The result was visually inspected and, if necessary, corrected for a few mistakenly detected clusters with only two circles. All fully depicted clusters >2 were analyzed for their major and minor axis length, as well as for their aspect ratio, with the “Measure” tool.
For the quantitative analysis of ORAI1 distribution in puncta, all recorded images from all maximally SOCE-activated cells were visually inspected in order to identify images displaying puncta. These puncta were defined as regions of locally accumulated labeled ORAI1 with at least two-fold higher label densities than in the surrounding regions, limited by a visually identifiable border towards the surrounding regions of lower ORAI1 density. Accumulated ORAI1 areas of strand- or ring-like shapes were excluded from the analysis, due to the difficulty of defining their borders. Of all recorded images from the group of maximally SOCE activated cells, 58% contained such puncta. With the “Freehand selection” tool in ImageJ, all fully depicted puncta were manually marked at their borders, thus yielding regions of interest (ROIs). The major and minor axis length, as well as their area size of these ROIs, was determined with the “Measure” tool. Thereafter, the labels inside the ROIs were blanked by filling the ROIs with grey background color, and the remaining number of labels in the regions outside the ROIs was again determined using the “Find particles” tool, similarly as done previously for all recorded images (as explained in the previous paragraph). The difference between the number of particles detected outside the ROI and the total number of particles in the respective image yielded the number of particles within the ROI.
4.8. Simulation of Label Distributions Corresponding to Selected STEM Image Data Sets
Spatial label distributions were simulated to discern if the detected label clusters >2 were bound to supramolecular ORAI1 arrangements, consisting of more than one underlying ORAI1 hexamer, or if similar clusters could also derive from randomly distributed hexameric ORAI1. These simulations were based on data recorded from different experimental groups, each consisting of a set of 12 images from cells displaying low ORAI1 expression levels, and a set of six images from cells with high expression levels. For every image, a corresponding simulation was made using the same number of detected particles and the image size but the distribution of the labels was determined by an algorithm based on randomly scattered hexameric ORAI1, labeled with a 30% labeling efficiency, and including effects of spatial hindrance of a newly bound QD to already bound QDs using a model of hard shell QD cores with a surrounding soft shell composed of the coating polymer and streptavidins. These simulated label distributions were processed and analyzed in the same way as the data from the STEM images.
When creating a new particle simulation, as a first step, a given number of virtual ORAI1 proteins were placed randomly within the image. Their positions followed a uniform distribution while overlapping proteins were strictly avoided by limiting the minimum distance between hexamers to 10 nm, representing the largest diameter of the protein complex located intracellularly. Each of the proteins was then loaded with up to six circular labels. The exact number of labels at each protein was determined by a random process modeling of the labeling efficiency. Also, the center-to-center distance between protein and label was generated randomly but restricted to a maximum of 20 nm. The directions of the displacements were generated randomly. Overlap of the hard shell of the labels (5 nm radius) was prohibited, that is, the minimal distance between two label centers was 10 nm.
Additionally, the soft shell of the labels (additional 3.5 nm radius) was modeled by sometimes allowing and sometimes prohibiting an overlap. The probability for accepting a particle’s position when soft shells were overlapping was calculated as:
with soft shell thickness
l = 3.5 nm, and the randomly generated distance between the hard shells
s ϵ [0, 2 × l]. This generated minimal acceptance probability if an overlap of the entire soft shell occurred, that is, the hard shells touched
. For soft shells that were barely touching (
s = 2 ×
l), the probability was maximized.
If a generated label position led to an overlap, as described above, with a previously placed label, a new position was created. If necessary, this was repeated multiple times before the label was discarded entirely, assuming that no valid position would be found in this area. The maximum number of attempts was 50. To avoid any bias, the labels were placed sequentially, starting with the first label of every single protein before continuing with the next ones. Finally, a simulated image was created from these label positions using the previously mentioned ImageJ plugin.
4.9. Statistical Analyses
Statistical significance was performed using the paired Student’s t-test. Differences were considered significant at p < 0.05 and are indicated as follows: * p < 0.05, ** p < 0.01 and *** p < 0.001. All values are expressed as average values with their standard deviation.