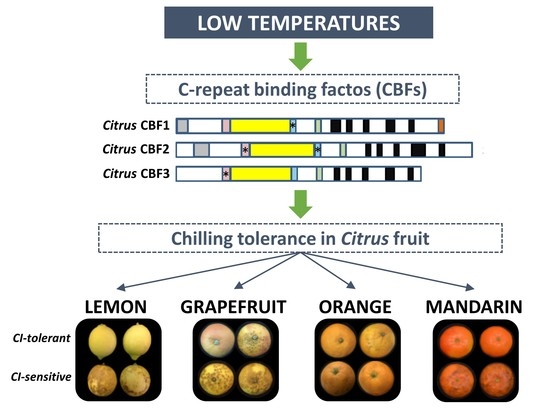
Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage
Abstract
:1. Introduction
2. Results
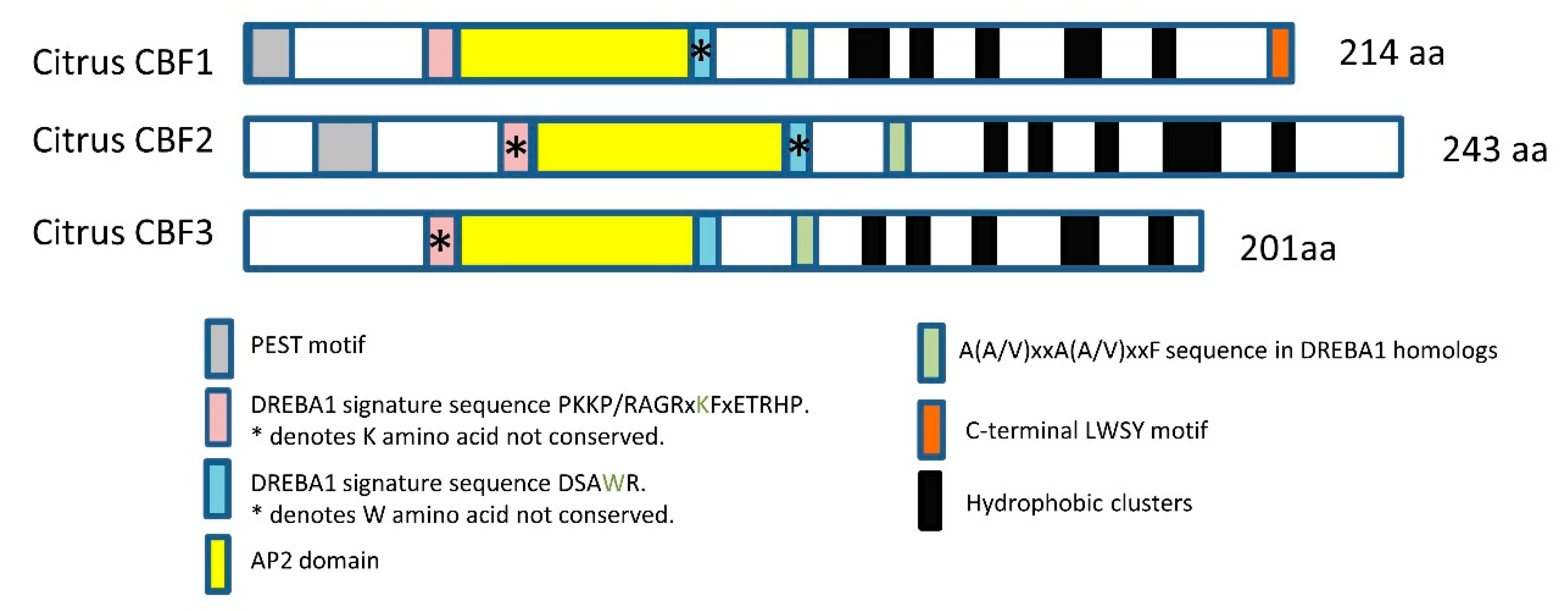
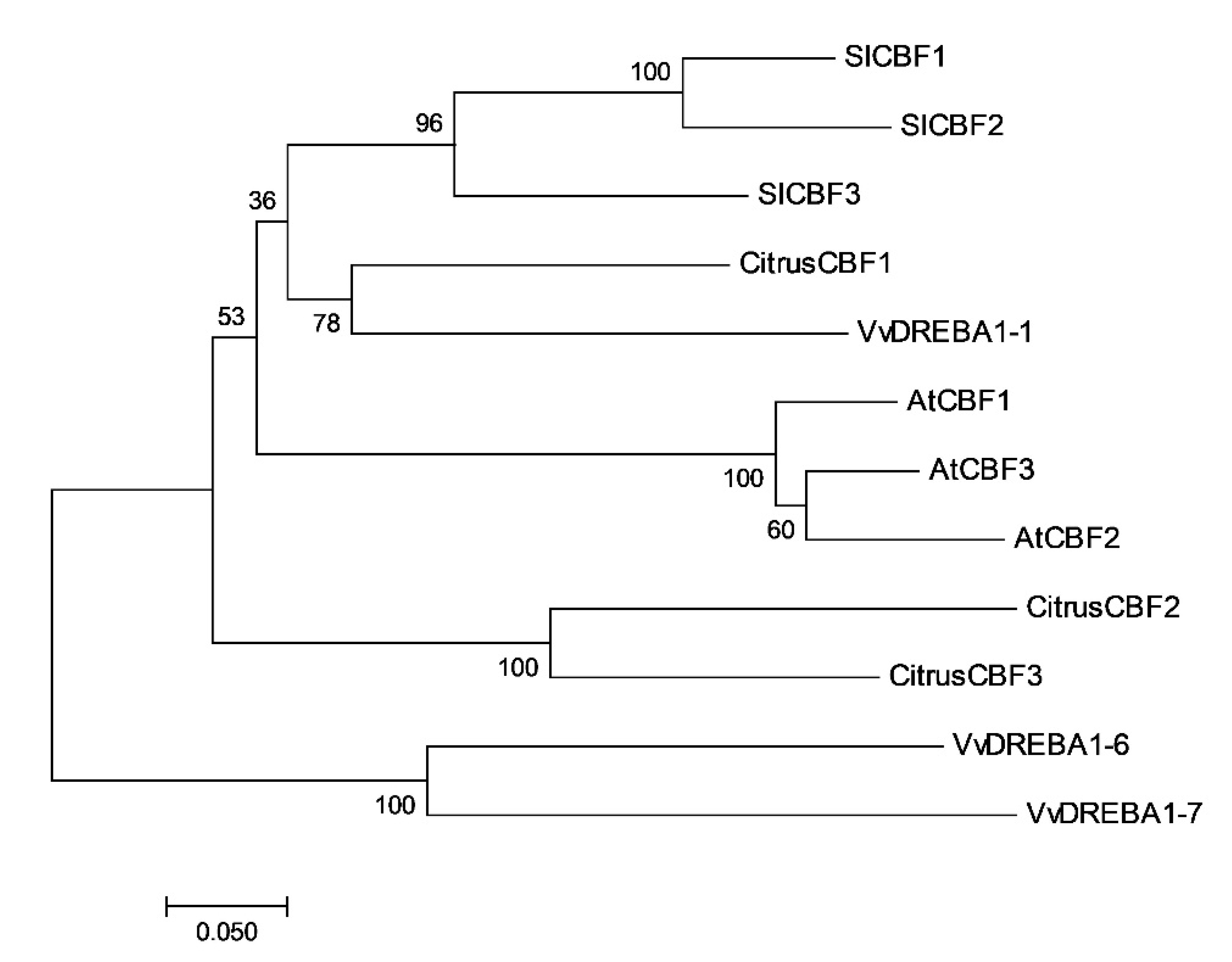
2.1. In Silico Study of Citrus CBFs
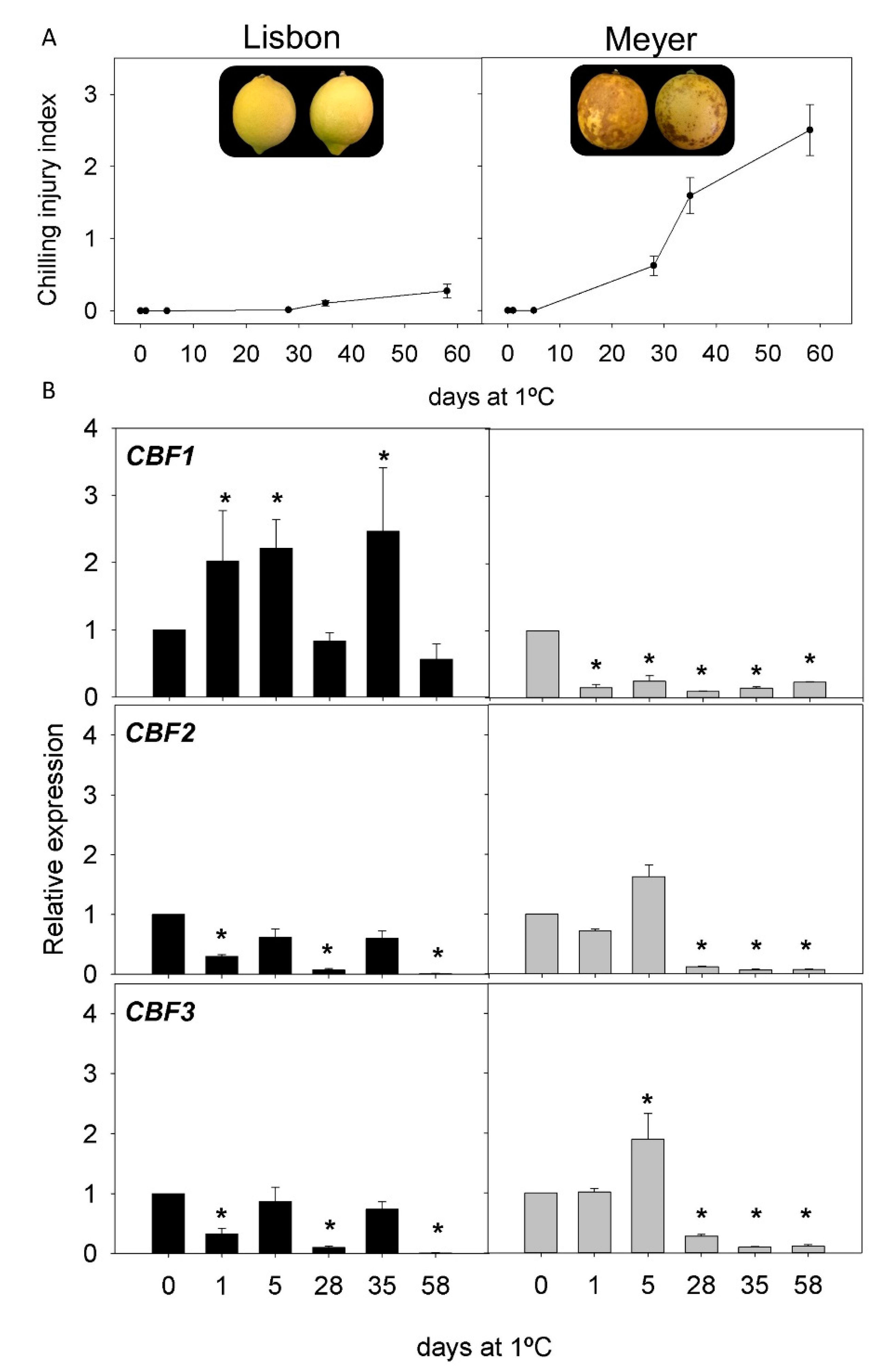
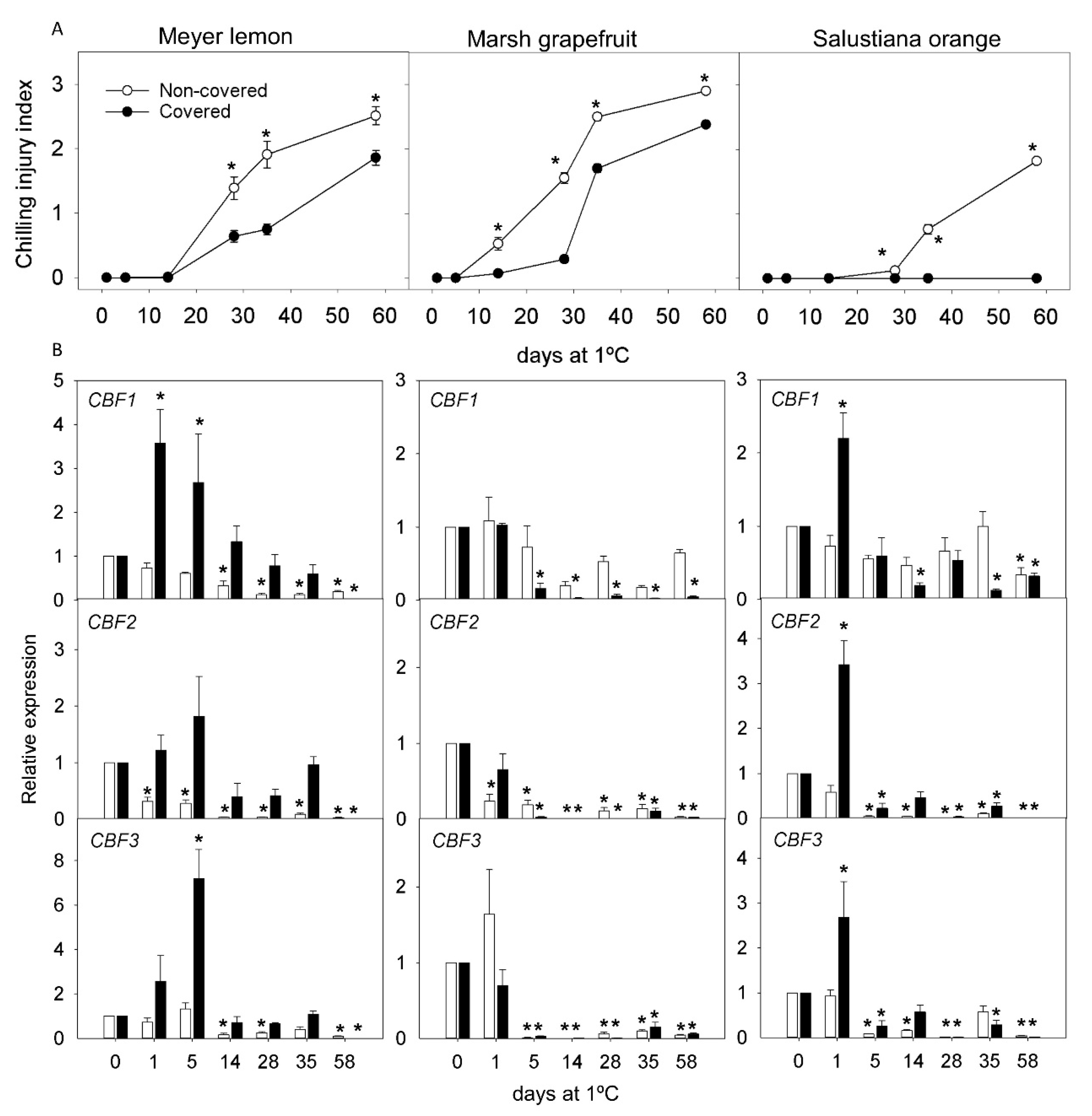
2.2. CI Symptoms and Expression of CBFs Genes in Cold-Tolerant and Cold-Sensitive Citrus Fruits during Cold Storage
2.3. Effect of Light Deprivation on CI and Expression of CBF Genes in Fruit of Cold-Sensitive Citrus Fruits during Cold Storage
3. Discussion
4. Materials and Methods
4.1. Plant Material, Preharvest Treatments, and Storage Conditions
4.2. Fruit Color and Chilling Injury Evaluation
4.3. RNA Extraction and Quantitative Real-Time PCR Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AP2 | Apetala2 |
| CI | Chilling injury |
| CBFs | C-repeat binding factors |
| COR genes | Cold regulated genes |
| ICC | Citrus color index |
| phyB | Phytochrome B |
| PIF3 | Phytochrome-interacting transcription 3 |
| DREB | Dehydration-responsive element binding |
| HCA | Hydrophobic cluster analysis |
References
- Biolatto, A.; Vazquez, D.E.; Sancho, A.M.; Carduza, F.J.; Pensel, N.A. Effect of commercial conditioning and cold quarantine storage treatments on fruit quality of “Rouge La Toma” grapefruit (Citrus paradisi Macf.). Postharvest Biol. Technol. 2005, 35, 167–176. [Google Scholar] [CrossRef]
- Lado, J.; Cronje, P.J.; Rodrigo, M.J.; Zacarías, L. Citrus. In Postharvest Physiological Disorders in Fruits and Vegetables; de Freitas, S.T., Sunil, P., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis: Abingdon, UK, 2019; pp. 377–398. ISBN 9781315267470. [Google Scholar]
- Lafuente, M.T.; Zacarías, L. Postharvest physiological disorders in citrus fruit. Stewart Postharvest Rev. 2006, 2, 1–9. [Google Scholar] [CrossRef]
- Dou, H. The influence of harvesting time and geographical location on susceptibility to physiological peel disorders associated with four Florida grapefruit cultivars. J. Hortic. Sci. Biotechnol. 2005, 80, 399–402. [Google Scholar] [CrossRef]
- Chalutz, E.; Waks, J.; Schiffmann-Nadel, M. A comparison of the response of different citrus fruit cultivars to storage temperature. Sci. Hortic. (Amsterdam) 1985, 25, 271–277. [Google Scholar] [CrossRef]
- Purvis, A.C. Relationship between mid-season resistance to chilling injury and reducing sugar level in grapefruit peel. HortScience 1979, 14, 227–229. [Google Scholar]
- Schirra, M.; Agabbio, M.; D’Hallewin, G. Chilling responses of grapefruit as affected by cultivar and harvest date. Adv. Hortic. Sci. 1998, 12, 118–122. [Google Scholar]
- Lafuente, M.T.; Martínez-Téllez, M.A.; Zacarías, L. Abscisic Acid in the Response of ‘Fortune’ Mandarins to Chilling. Effect of Maturity and High-Temperature Conditioning. J. Sci. Food Agric. 1997, 73, 494–502. [Google Scholar] [CrossRef]
- Lado, J.; Cronje, P.; Alquézar, B.; Page, A.; Manzi, M.; Gómez-Cadenas, A.; Stead, A.D.; Zacarías, L.; Rodrigo, M.J. Fruit shading enhances peel color, carotenes accumulation and chromoplast differentiation in red grapefruit. Physiol. Plant. 2015, 154, 469–484. [Google Scholar] [CrossRef] [Green Version]
- Lado, J.; Rodrigo, M.J.; Cronje, P.; Zacarías, L. Involvement of lycopene in the induction of tolerance to chilling injury in grapefruit. Postharvest Biol. Technol. 2015, 100, 176–186. [Google Scholar] [CrossRef]
- Lado, J.; Rodrigo, M.J.; López-Climent, M.; Gómez-Cadenas, A.; Zacarías, L. Implication of the antioxidant system in chilling injury tolerance in the red peel of grapefruit. Postharvest Biol. Technol. 2016, 111, 214–223. [Google Scholar] [CrossRef]
- Sapitnitskaya, M.; Maul, P.; McCollum, G.T.; Guy, C.L.; Weiss, B.; Samach, A.; Porat, R. Postharvest heat and conditioning treatments activate different molecular responses and reduce chilling injuries in grapefruit. J. Exp. Bot. 2006, 57, 2943–2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, A.; Li, W.; Ye, J.; Sun, X.; Ding, Y.; Cheng, Y.; Deng, X. Microarray Expression Profiling of Postharvest Ponkan Mandarin (Citrus reticulata) Fruit under Cold Storage Reveals Regulatory Gene Candidates and Implications on Soluble Sugars Metabolism. J. Integr. Plant. Biol. 2011, 53, 358–374. [Google Scholar] [CrossRef] [PubMed]
- Maul, P.; McCollum, G.T.; Popp, M.; Guy, C.L.; Porat, R. Transcriptome profiling of grapefruit flavedo following exposure to low temperature and conditioning treatments uncovers principal molecular components involved in chilling tolerance and susceptibility. Plant Cell Environ. 2008, 31, 752–768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.Q.; Shen, C.; Wu, L.H.; Tang, K.X.; Lin, J. CBF-dependent signaling pathway: A key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 2011, 31, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Champ, K.I.; Febres, V.J.; Moore, G.A. The role of CBF transcriptional activators in two Citrus species (Poncirus and Citrus) with contrasting levels of freezing tolerance. Physiol. Plant 2007, 129, 529–541. [Google Scholar] [CrossRef]
- He, L.G.; Wang, H.L.; Liu, D.C.; Zhao, Y.J.; Xu, M.; Zhu, M.; Wei, G.Q.; Sun, Z.H. Isolation and expression of a cold-responsive gene PtCBF in Poncirus trifoliata and isolation of citrus CBF promoters. Biol. Plant. 2012, 56, 484–492. [Google Scholar] [CrossRef]
- He, L.; Jiang, Y.; Wang, H.; Xu, M.; Sun, Z. Expression and regulation of a cold-responsive gene, CsCBF in Citrus sinensis (L.) Osbeck under low temperature, high salinity and abscisic acid. Acta Hortic. 2016, 1135, 33–46. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. In Pllant Stress Tolerance. Methods in Molecular Biology; Sunkar, R., Ed.; Humana Press: Totowa, NJ, USA, 2010; Volume 639, pp. 39–55. ISBN 978-1-60761-701-3. [Google Scholar]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant. J. 2004, 39, 905–919. [Google Scholar] [CrossRef]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant. Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi, M.; Abdullah, S.N.A.; Aziz, M.A.; Namasivayam, P. A novel CBF that regulates abiotic stress response and the ripening process in oil palm (Elaeis guineensis) fruits. Tree Genet. Genomes 2015, 11. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.-Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant. Cell Physiol. 2004, 45, 1042–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate Regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 Cascade and Freezing Tolerance in Arabidopsis. Plant. Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Zhang, B.; Yin, X.-R.; Xu, C.-J.; Sun, C.-D.; Chen, K.-S. Differential Expression of the CBF Gene Family During Postharvest Cold Storage and Subsequent Shelf-Life of Peach Fruit. Plant Mol. Biol. Rep. 2013, 31, 1358–1367. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia). Int. J. Mol. Sci. 2019, 20, 2074. [Google Scholar] [CrossRef] [Green Version]
- Şahin-Çevik, M.; Moore, G.A. Two AP2 domain containing genes isolated from the cold-hardy Citrus relative Poncirus trifoliata are induced in response to cold. Funct. Plant Biol. 2006, 33, 863. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, J.; Sun, T.; Wang, X.; Zhu, C.; Ai, Y.; Gu, H. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-J.; Kim, Y.-K.; Park, J.-Y.; Kim, J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002, 29, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-induced CBF–PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2019, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Shen, L.; Fan, B.; Yu, M.; Zheng, Y.; Lv, S.; Sheng, J. Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits. FEBS Lett. 2009, 583, 3329–3334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Zhang, Q.; Pan, Y.; Che, F.; Wang, Q.; Meng, X.; Rao, J. Changes of polyamines and CBFs expressions of two Hami melon (Cucumis melo L.) cultivars during low temperature storage. Sci. Hortic. (Amsterdam) 2017, 224, 8–16. [Google Scholar] [CrossRef]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Hernandez, M.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. Deciphering the Role of CBF/DREB Transcription Factors and Dehydrins in Maintaining the Quality of Table Grapes cv. Autumn Royal Treated with High CO2 Levels and Stored at 0 °C. Front. Plant Sci. 2017, 8, 1591. [Google Scholar] [CrossRef]
- Fernandez-Caballero, C.; Rosales, R.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. Unraveling the roles of CBF1, CBF4 and dehydrin 1 genes in the response of table grapes to high CO2 levels and low temperature. J. Plant Physiol. 2012, 169, 744–748. [Google Scholar] [CrossRef] [Green Version]
- Haake, V.; Cook, D.; Riechmann, L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription Factor CBF4 Is a Regulator of Drought Adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Gregorio, J.; Hernández-bernal, A.F.; Cordoba, E.; León, P. Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA-INSENSITIVE (ABI) 4 transcription factor. Mol. Plant 2014, 7, 422–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, M.; Jaiswal, A.; Taj, G.; Jaiswal, J.P.; Qureshi, M.I.; Singh, N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012, 91, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Fei, S. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 2006, 224, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Triezenberg, S.J.; Thomashow, M.F.; Stockinger, E.J. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol. Biol. 2005, 543–559. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution (N. Y). 1985, 39, 783. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Costa-Broseta, Á.; Perea-Resa, C.; Castillo, M.; Salinas, J.; León, J. Nitric oxide deficiency decreases C-repeat binding factor-dependent and -independent induction of cold acclimation. J. Exp. Bot. 2019, 3285–3296. [Google Scholar] [CrossRef] [Green Version]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Yahia, N.; Wani, S.H.; Kumar, V. CBF-Dependent and CBF-Independent transcriptional regulation of cold stress responses in plants. In Cold Tolerance in Plants; Wani, S.H., Herath, V., Eds.; Springer: Cham, Switzerland, 2018; pp. 89–102. ISBN 9783030014155. [Google Scholar]
- Pons, C.; Martí, C.; Forment, J.; Crisosto, C.H.; Dandekar, A.M.; Granell, A. A bulk segregant gene expression analysis of a peach population reveals components of the underlying mechanism of the fruit cold response. PLoS ONE 2014, 9, e90706. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Q.; Hu, M.; Gao, Z.; An, F.; Li, M.; Jiang, Y. Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food Chem. 2017, 219, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Sheng, J.; Lv, S.; Zheng, Y.; Zhang, J.; Yu, M.; Shen, L. Nitric oxide participates in the regulation of LeCBF1 gene expression and improves cold tolerance in harvested tomato fruit. Postharvest Biol. Technol. 2011, 62, 121–126. [Google Scholar] [CrossRef]
- Albornoz, K.; Cantwell, M.I.; Zhang, L.; Beckles, D.M. Integrative analysis of postharvest chilling injury in cherry tomato fruit reveals contrapuntal spatio- temporal responses to ripening and cold stress. Sci. Rep. 2019, 9–2795, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacarias, L.; Cronje, P.J.R.; Palou, L. Postharvest technology of citrus fruits. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 421–446. ISBN 978-0-12-812163-4. [Google Scholar]
- Curk, F.; Navarro, L. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canella, D.; Gilmour, S.J.; Kuhn, L.A.; Thomashow, M.F. Biochimica et Biophysica Acta DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 2010, 1799, 454–462. [Google Scholar] [CrossRef]
- Lado, J.; Alós, E.; Rodrigo, M.J.; Zacarías, L. Light avoidance reduces ascorbic acid accumulation in the peel of Citrus fruit. Plant Sci. 2015, 231, 138–147. [Google Scholar] [CrossRef]
- Magwaza, L.S.; Opara, U.L.; Cronje, P.J.R.; Landahl, S.; Terry, L.A. Canopy position affects rind biochemical profile of “Nules Clementine” mandarin fruit during postharvest storage. Postharvest Biol. Technol. 2013, 86, 300–308. [Google Scholar] [CrossRef]
- Lado, J.; Alós, E.; Manzi, M.; Cronje, P.J.R.; Gómez-Cadenas, A.; Rodrigo, M.J.; Zacarías, L. Light Regulation of Carotenoid Biosynthesis in the Peel of Mandarin and Sweet Orange Fruits. Front. Plant Sci. 2019, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Differential transcriptional regulation of l-ascorbic acid content in peel and pulp of citrus fruits during development and maturation. Planta 2014, 239, 1113–1128. [Google Scholar] [CrossRef] [PubMed]




| Non covered | Covered | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Specie | Variety | CI | CBF1 | CBF2 | CBF3 | CI | CBF1 | CBF2 | CBF3 |
| Lemon | Lisbon | - | ++ | - | - | n.d. | |||
| Meyer | +++ | -- | + | + | + | ++ | - | +++ | |
| Grapefruit | Star Ruby | + | +++ | + | + | n.d. | |||
| Marsh | +++ | -- | - | - | ++ | - | - | - | |
| Orange | Navelina | - | +++ | - | - | n.d. | |||
| Salustiana | ++ | - | - | - | -- | + | ++ | ++ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvo, M.; Rey, F.; Arruabarrena, A.; Gambetta, G.; Rodrigo, M.J.; Zacarías, L.; Lado, J. Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage. Int. J. Mol. Sci. 2021, 22, 804. https://doi.org/10.3390/ijms22020804
Salvo M, Rey F, Arruabarrena A, Gambetta G, Rodrigo MJ, Zacarías L, Lado J. Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage. International Journal of Molecular Sciences. 2021; 22(2):804. https://doi.org/10.3390/ijms22020804
Chicago/Turabian StyleSalvo, Matías, Florencia Rey, Ana Arruabarrena, Giuliana Gambetta, María J. Rodrigo, Lorenzo Zacarías, and Joanna Lado. 2021. "Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage" International Journal of Molecular Sciences 22, no. 2: 804. https://doi.org/10.3390/ijms22020804
APA StyleSalvo, M., Rey, F., Arruabarrena, A., Gambetta, G., Rodrigo, M. J., Zacarías, L., & Lado, J. (2021). Transcriptional Analysis of C-Repeat Binding Factors in Fruit of Citrus Species with Differential Sensitivity to Chilling Injury during Postharvest Storage. International Journal of Molecular Sciences, 22(2), 804. https://doi.org/10.3390/ijms22020804