Cytological Observations and Bulked-Segregant Analysis Coupled Global Genome Sequencing Reveal Two Genes Associated with Pollen Fertility in Tetraploid Rice
Abstract
:1. Introduction
2. Results
2.1. Morphological and Cytological Observations of Bulks (JG and JD) with Different Fertility
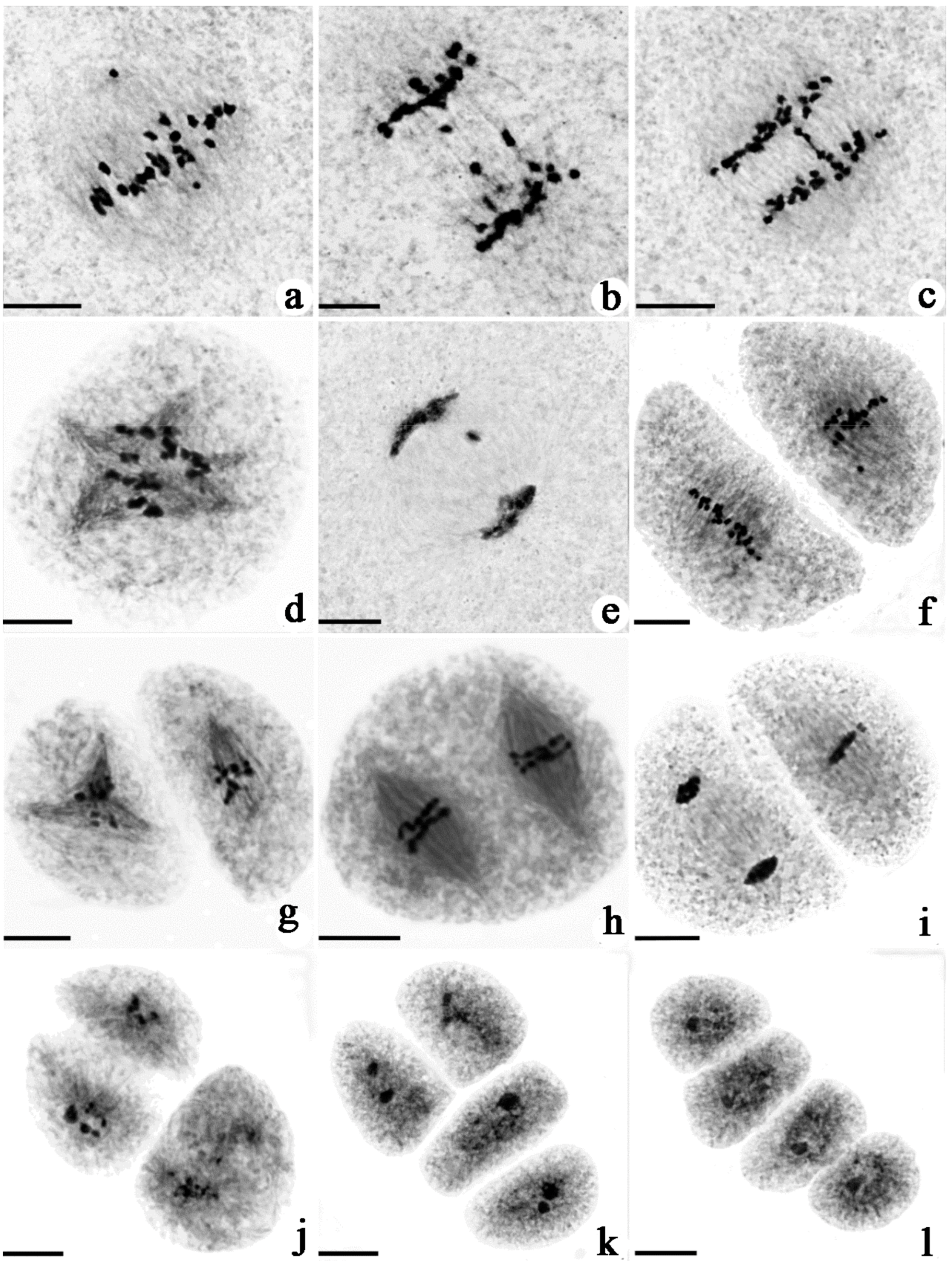
2.2. Comparison of Chromosomal Behavior between H3, T452, JG and JD
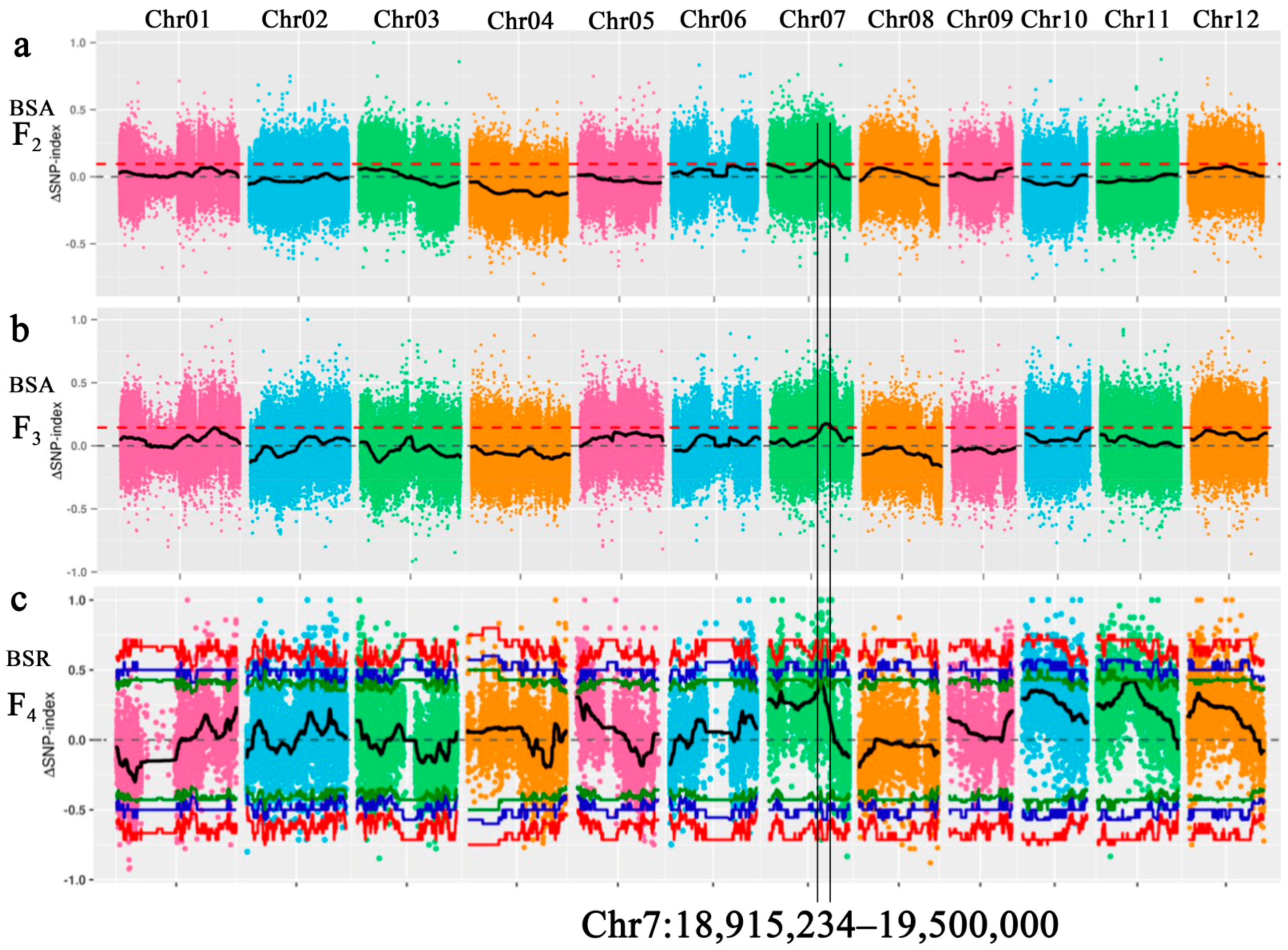
2.3. High Fertility Blocks Identification by BSA-seq and BSR-seq Using JG Bulk, JD Bulk and Their Parents
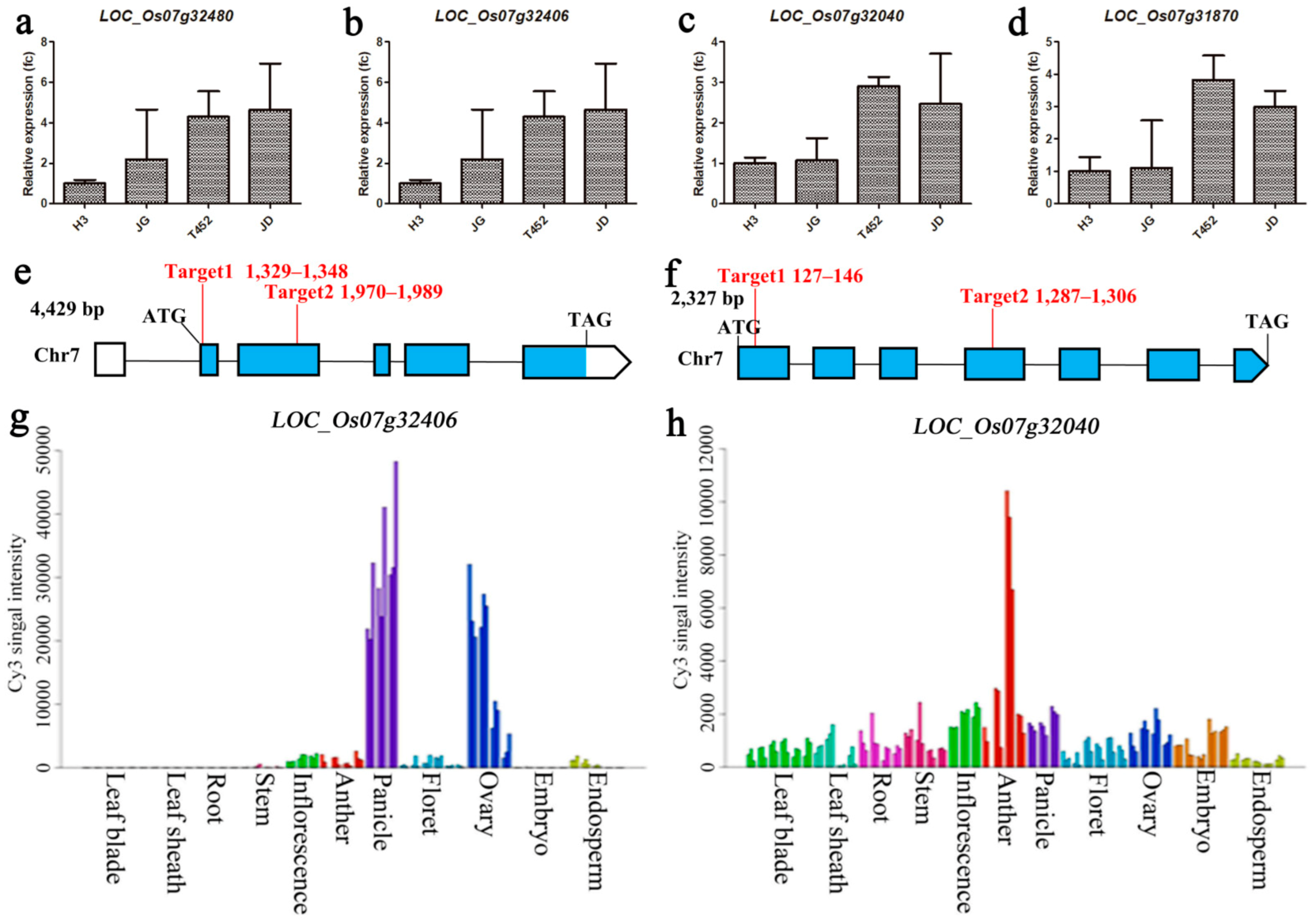
2.4. Expression Analysis of Candidate Genes in High Fertility Blocks
2.5. Knockout of Two Candidate Genes Causes Pollen Abortion in Neo-Tetraploid Rice
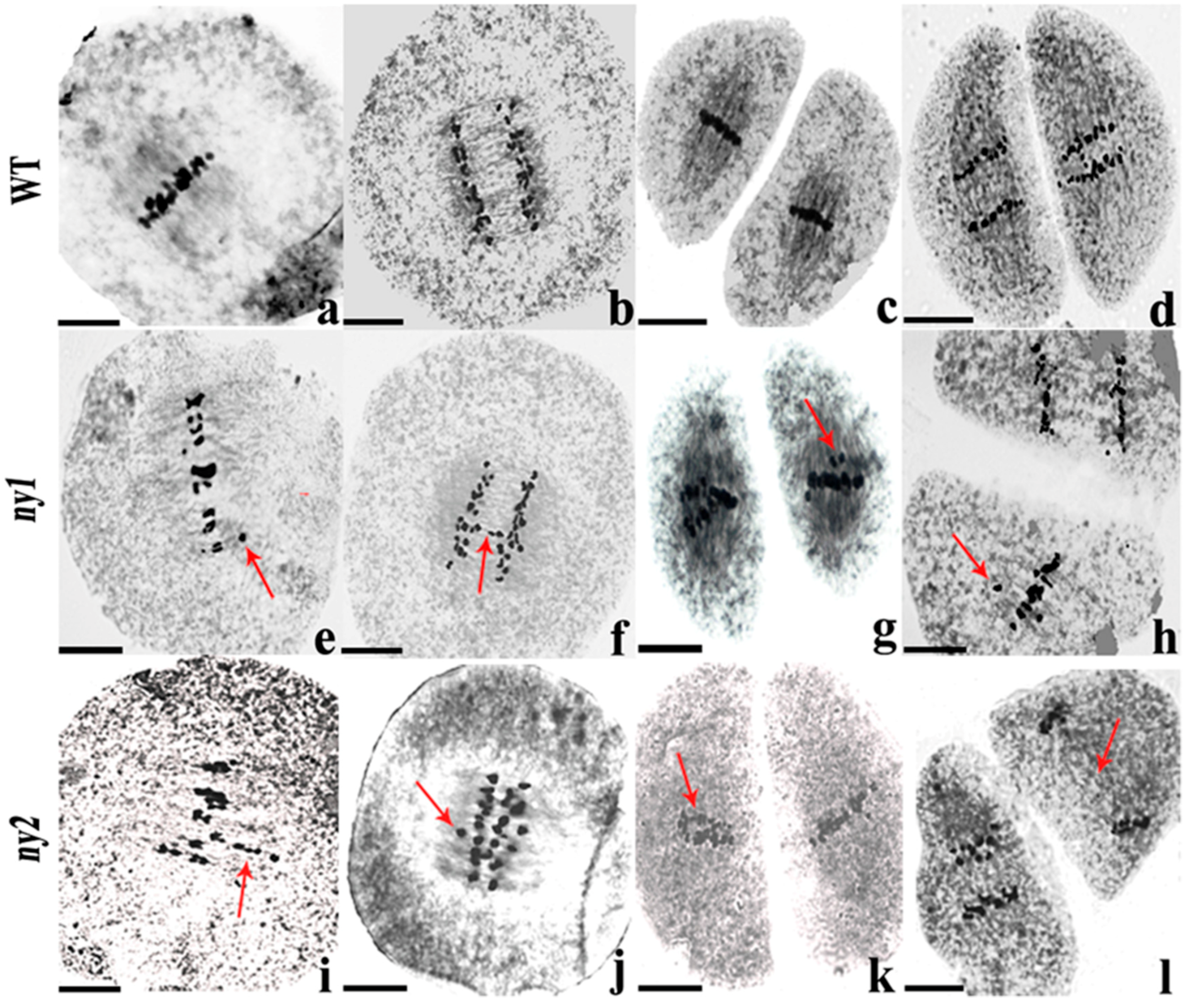
2.6. Analysis of Chromosome Behavior in ny1, ny2 and WT
3. Discussion
3.1. BSA-Seq is an Effective Technique to Identify Genes in Polyploid Plants
3.2. NY1 and NY2 Play Crucial Role in Pollen Mother Cell Meiosis
4. Materials and Methods
4.1. Plant Material
4.2. Cytological Observation
4.3. qRT-PCR Analysis of Candidate Gene Expression
4.4. Mixed Pool Construction and Analysis of Mixed Pool Sequencing Data
4.5. Development and Identification of Mutant Plants in Huaduo1
4.6. Investigation of Agronomic Traits and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | Bulked segregant analysis |
| DAF | Days after flowering |
| DGEs | Differentially expressed genes |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| PMCs | Pollen mother cells |
| QTL | Quantitative trait locus |
| WE-CLSM | Whole-mount eosin B-staining confocal laser scanning microscopy |
| WT | Wild type |
References
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Valkonen, J.P. Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 2001, 39, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Van De Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; De Pamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shahid, M.Q.; Guo, H.; Yin, W.; Chen, Z.; Wang, L.; Liu, X.-D.; Lu, Y.-G. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 2014, 27, 181–196. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Liu, G.; Li, J.; Naeem, M.; Liu, X.-D. Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agric. Scand. Sect. B: Plant Soil Sci. 2011, 61, 23–32. [Google Scholar] [CrossRef]
- Shahid, M.Q.; Xu, H.; Lin, S.; Chen, Z.; Naeem, M.; Li, Y.; Liu, X. Genetic analysis and hybrid vigor study of grain yield and other quantitative traits in autotetraploid rice. Pak. J. Bot. 2012, 44, 237–246. [Google Scholar]
- Guo, H.; Mendrikahy, J.N.; Xie, L.; Deng, J.; Lu, Z.; Wu, J.; Li, X.; Shahid, M.Q.; Liu, X. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 2017, 7, 40139. [Google Scholar] [CrossRef] [Green Version]
- Koide, Y.; Kuniyoshi, D.; Kishima, Y. Fertile Tetraploids: New Resources for Future Rice Breeding? Front. Plant Sci. 2020, 11, 1231. [Google Scholar] [CrossRef]
- Wu, J.; Shahid, M.Q.; Chen, L.; Chen, Z.; Wang, L.; Liu, X.-D.; Lu, Y. Polyploidy Enhances F1 Pollen Sterility Loci Interactions That Increase Meiosis Abnormalities and Pollen Sterility in Autotetraploid Rice. Plant Physiol. 2015, 169, 2700–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, M.Q.; Li, Y.; Saleem, M.F.; Naeem, M.; Wei, C.; Liu, X. Yield and yield components in autotetraploid and diploid rice genotypes (indica and japonica) sown in early and late seasons. Aust. J. Crop Sci. 2013, 7, 632–641. [Google Scholar]
- Li, X.; Shahid, M.Q.; Xiang, L.; Lu, Z.; Fang, N.; Wang, L.; Wu, J.; Chen, Z.; Liu, X.-D. Analysis of small RNAs revealed differential expressions during pollen and embryo sac development in autotetraploid rice. BMC Genom. 2017, 18, 129. [Google Scholar] [CrossRef] [Green Version]
- Shahid, M.Q.; Sun, J.; Wei, C.; Zhang, P.; Liu, X. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 2010, 42, 7–19. [Google Scholar]
- Cai, D.; Chen, J.; Chen, D.; Dai, B.; Zhang, W.; Song, Z.; Yang, Z.; Du, C.; Tang, Z.; He, Y.; et al. The breeding of two polyploid rice lines with the characteristic of polyploid meiosis stability. Sci. China Ser. C Life Sci. 2007, 50, 356–366. [Google Scholar] [CrossRef]
- Ghaleb, M.A.A.; Li, C.; Shahid, M.Q.; Yu, H.; Liang, J.; Chen, R.; Wu, J.; Liu, X.-D. Heterosis analysis and underlying molecular regulatory mechanism in a wide-compatible neo-tetraploid rice line with long panicles. BMC Plant Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yuan, Y.; Wu, J.; Chen, Z.; Wang, L.; Shahid, M.Q.; Liu, X.-D. Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice 2019, 12, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Bei, X.; Shahid, M.Q.; Wu, J.; Chen, Z.; Wang, L.; Liu, X.-D. Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice. PLoS ONE 2019, 14, e0214953. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yu, H.; Jiao, Y.; Shahid, M.Q.; Wu, J.; Liu, X.-D. Genome-wide analysis of DNA polymorphisms, the methylome and transcriptome revealed that multiple factors are associated with low pollen fertility in autotetraploid rice. PLoS ONE 2018, 13, e0201854. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.; Lin, H.; Chen, Y.; Yu, H.; Lu, Z.; Li, X.; Zhou, H.; Chen, Z.; Liu, X.-D. Comparative Cytological and Transcriptome Analysis Revealed the Normal Pollen Development Process and up-Regulation of Fertility-Related Genes in Newly Developed Tetraploid Rice. Int. J. Mol. Sci. 2020, 21, 7046. [Google Scholar] [CrossRef]
- Lu, Z.; Guo, X.; Huang, Z.; Xia, J.; Li, X.; Wu, J.; Yu, H.; Shahid, M.Q.; Liu, X.-D. Transcriptome and Gene Editing Analyses Reveal MOF1a Defect Alters the Expression of Genes Associated with Tapetum Development and Chromosome Behavior at Meiosis Stage Resulting in Low Pollen Fertility of Tetraploid Rice. Int. J. Mol. Sci. 2020, 21, 7489. [Google Scholar] [CrossRef]
- Xiong, Y.; Gan, L.; Hu, Y.; Sun, W.; Zhou, X.; Song, Z.; Zhang, X.; Li, Y.; Yang, Z.; Xu, W.; et al. OsMND1 regulates early meiosis and improves the seed set rate in polyploid rice. Plant Growth Regul. 2019, 87, 341–356. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [Green Version]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Yan, W.; Chen, Z.; Lu, J.; Xu, C.; Xie, G.; Li, Y.; Deng, X.W.; He, H.; Tang, X. Simultaneous Identification of Multiple Causal Mutations in Rice. Front. Plant Sci. 2017, 7, 2055. [Google Scholar] [CrossRef] [Green Version]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap+: Genetic Mapping and Mutant Identification without Crossing in Rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Zhao, Y.; Zhu, Q.; Zhang, Z.; Fan, C.; Amanullah, S.; Gao, P.; Luan, F. Mapping of powdery mildew resistance genes in melon (Cucumis melo L.) by bulked segregant analysis. Sci. Hortic. 2017, 220, 160–167. [Google Scholar] [CrossRef]
- Song, J.; Li, Z.; Liu, Z.; Guo, Y.; Qiu, L.-J. Next-Generation Sequencing from Bulked-Segregant Analysis Accelerates the Simultaneous Identification of Two Qualitative Genes in Soybean. Front. Plant Sci. 2017, 8, 919. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Sun, S.; Li, Y.; Duan, C.; Zhu, Z. Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel Phytophthora resistance gene, RpsHC18, in soybean. Theor. Appl. Genet. 2017, 131, 525–538. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, T.; You, X.; Jiang, J.; Li, J.; Xu, X. Molecular mapping of the Cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato (Solanum lycopersicum). BMC Plant Biol. 2019, 19, 15. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Singh, M.; Srivastava, R.; Bajaj, D.; Saxena, M.S.; Rana, J.C.; Bansal, K.C.; Tyagi, A.K.; Parida, S.K. mQTL-seq delineates functionally relevant candidate gene harbouring a major QTL regulating pod number in chickpea. DNA Res. 2015, 23, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.K.; Khan, A.W.; Jaganathan, D.; Thudi, M.; Roorkiwal, M.; Takagi, H.; Garg, V.; Kumar, V.; Chitikineni, A.; Gaur, P.M.; et al. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 2016, 14, 2110–2119. [Google Scholar] [CrossRef]
- Liang, D.; Chen, M.; Qi, X.; Xu, Q.; Zhou, F.; Chen, X. QTL Mapping by SLAF-seq and Expression Analysis of Candidate Genes for Aphid Resistance in Cucumber. Front. Plant Sci. 2016, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wang, H.; Huang, X.; Wang, Q.; Wang, J.; An, D.; Li, J.; Wang, W.; Wu, Y. Maize VKS1 Regulates Mitosis and Cytokinesis During Early Endosperm Development. Plant Cell 2019, 31, 1238–1256. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xiao, L.; Guo, S.; An, F.; Du, D. Fine Mapping and Whole-Genome Resequencing Identify the Seed Coat Color Gene in Brassica rapa. PLoS ONE 2016, 11, e0166464. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2006, 35, D883–D887. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Antonio, B.A.; Namiki, N.; Takehisa, H.; Minami, H.; Kamatsuki, K.; Sugimoto, K.; Shimizu, Y.; Hirochika, H.; Nagamura, Y. RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2010, 39, D1141–D1148. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Zhang, X.; Li, R.; Chen, L.; Niu, S.; Chen, L.; Gao, J.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; et al. Fine-mapping and candidate gene analysis of the Brassica juncea white-flowered mutant Bjpc2 using the whole-genome resequencing. Mol. Genet Genom. 2018, 293, 359–370. [Google Scholar] [CrossRef]
- Luo, Q.; Li, Y.; Shen, Y.; Cheng, Z. Ten Years of Gene Discovery for Meiotic Event Control in Rice. J. Genet. Genom. 2014, 41, 125–137. [Google Scholar] [CrossRef] [Green Version]
- Kuniyoshi, D.; Masuda, I.; Kanaoka, Y.; Shimazaki-Kishi, Y.; Okamoto, Y.; Yasui, H.; Yamamoto, T.; Nagaki, K.; Hoshino, Y.; Koide, Y.; et al. Diploid Male Gametes Circumvent Hybrid Sterility Between Asian and African Rice Species. Front. Plant Sci. 2020, 11, 1–27. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Q.; Tang, D.; Liu, X.; Du, G.; Shen, Y.; Li, Y.; Cheng, Z. OsDMC1 Is not Required for Homologous Pairing in Rice Meiosis. Plant Physiol. 2016, 171, 230–241. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Tang, D.; Wang, M.; Li, Y.; Zhang, L.; Wang, K.; Li, M.; Cheng, Z. MRE11 is required for homologous synapsis and DSB processing in rice meiosis. Chromosoma 2013, 122, 363–376. [Google Scholar] [CrossRef]
- Wang, K.; Wang, M.; Tang, D.; Shen, Y.; Miao, C.; Hu, Q.; Lu, T.; Cheng, Z. The Role of Rice HEI10 in the Formation of Meiotic Crossovers. PLoS Genet. 2012, 8, e1002809. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Tang, D.; Wang, K.; Wang, M.; Huang, J.; Luo, W.; Luo, Q.; Hong, L.; Li, M.; Cheng, Z. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 2012, 125, 2581–2591. [Google Scholar] [CrossRef] [Green Version]
- Shao, T.; Tang, D.; Wang, K.; Wang, M.; Che, L.; Qin, B.; Yu, H.; Li, M.; Gu, M.; Cheng, Z. OsREC8 Is Essential for Chromatid Cohesion and Metaphase I Monopolar Orientation in Rice Meiosis. Plant Physiol. 2011, 156, 1386–1396. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tang, D.; Wang, K.; Shen, Y.; Qin, B.; Miao, C.; Li, M.; Cheng, Z. OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J. 2011, 67, 583–594. [Google Scholar] [CrossRef]
- Luan, L.; Tu, S.B.; Long, W.B.; Wang, X.; Liu, Y.H.; Kong, F.L.; He, T.; Yan, W.G.; Yu, M.Q. Cytogenetic studies on two F1 hybrids of autotetraploid rice varieties showing extremely high level of heterosis. Plant Syst. Evol. 2007, 267, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Luan, L.; Wang, X.; Long, W.B.; Liu, Y.H.; Tu, S.B.; Xiao, X.Y.; Kong, F.L. A comparative cytogenetic study of the rice (Oryza sativa L.) autotetraploid restorers and hybrids. Генетика 2009, 45, 1074–1081. [Google Scholar] [CrossRef]
- He, J.-H.; Shahid, M.Q.; Chen, Z.-X.; Chen, X.-A.; Liu, X.-D.; Lu, Y.-G. Abnormal PMC microtubule distribution pattern and chromosome behavior resulted in low pollen fertility of an intersubspecific autotetraploid rice hybrid. Plant Syst. Evol. 2011, 291, 257–265. [Google Scholar] [CrossRef]
- He, J.H.; Shahid, M.Q.; Li, Y.J.; Guo, H.B.; Cheng, X.A.; Liu, X.-D.; Lu, Y.-G. Allelic interaction of F1 pollen sterility loci and abnormal chromosome behaviour caused pollen sterility in intersubspecific autotetraploid rice hybrids. J. Exp. Bot. 2011, 62, 4433–4445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Shahid, M.Q.; Wu, J.; Chen, Z.; Wang, L.; Liu, X.-D. Cytological and transcriptome analyses reveal abrupt gene expression for meiosis and saccharide metabolisms that associated with pollen abortion in autotetraploid rice. Mol. Genet. Genom. 2018, 293, 1407–1420. [Google Scholar] [CrossRef] [Green Version]
- Koide, Y.; Ogino, A.; Yoshikawa, T.; Kitashima, Y.; Saito, N.; Kanaoka, Y.; Onishi, K.; Yoshitake, Y.; Tsukiyama, T.; Saito, H.; et al. Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc. Natl. Acad. Sci. USA 2018, 115, E1955–E1962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, M.; Horiuchi, Y.; Ueda, Y.; Mizuta, Y.; Kubo, T.; Yano, K.; Yamaki, S.; Tsuda, K.; Nagata, T.; Niihama, M.; et al. Rice Expression Atlas In Reproductive Development. Plant Cell Physiol. 2010, 51, 2060–2081. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zhang, Z.-Y.; Zhang, W.-J.; Zhao, X.-M.; Li, X.; Zhang, D.; Liu, Q.-Q.; Tang, W.-H. Global Gene Profiling of Laser-Captured Pollen Mother Cells Indicates Molecular Pathways and Gene Subfamilies Involved in Rice Meiosis. Plant Physiol. 2010, 154, 1855–1870. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tang, D.; Luo, Q.; Jin, Y.; Shen, Y.; Wang, K.; Cheng, Z. BRK1, a Bub1-Related Kinase, Is Essential for Generating Proper Tension between Homologous Kinetochores at Metaphase I of Rice Meiosis. Plant Cell 2012, 24, 4961–4973. [Google Scholar] [CrossRef] [Green Version]
- Ghouri, F.; Zhu, J.; Yu, H.; Wu, J.; Baloch, F.S.; Liu, X.-D.; Shahid, M.Q. Deciphering global DNA variations and embryo sac fertility in autotetraploid rice line. Turk. J. Agric. For. 2019, 43, 554–568. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Shahid, M.Q.; Li, R.; Li, W.; Liu, W.; Ghouri, F.; Liu, X.-D. Genome-Wide Analysis of Genetic Variations and the Detection of Rich Variants of NBS-LRR Encoding Genes in Common Wild Rice Lines. Plant Mol. Biol. Rep. 2018, 36, 618–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.B.; Daly, M.J.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.-C.; Yost, H.J. MMAPPR: Mutation Mapping Analysis Pipeline for Pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [Green Version]


| Traits | H3 | T452 | JG | JD |
|---|---|---|---|---|
| Total grains/panicle | 136.9 ± 13.4 B | 116.0 ± 25.0 C | 159.4 ± 22.2 D | 120.7 ± 46.1 BC |
| Seed setting (%) | 80.4 ± 3.0 A | 15.3 ± 4.7 C | 82.5 ± 4.2 A | 18.8 ± 9.8 B |
| Pollen fertility (%) | 87.5 ± 2.4 A | 23.7 ± 6.8 B | 88.5 ± 4.8 A | 25.1 ± 5.1 B |
| Embryo sac fertility (%) | 92.0 | 85.2 | 93.5 | 85.0 |
| Embryo sac fertility at 3 DAF (%) | 82.0 | 11.0 | 81.9 | 24.9 |
| Chromosome lagging at metaphase I (%) | 17.4 ± 1.7 | 13.9 ± 1.3 | 16.6 ± 2.2 | 31.3 ± 4.8 |
| Chromosome straggling at anaphase I (%) | 17.6 ± 0.3 | 13.6 ± 1.4 | 8.4 ± 0.6 | 28.8 ± 7.4 |
| Chromosome lagging at metaphase II (%) | 5.4 ± 0.3 | 3.1 ± 1.9 | 2.4 ± 1.4 | 27.2 ± 1.4 |
| Chromosome straggling at anaphase II (%) | 0.7 ± 0.7 | 0.0 ± 0.0 | 2.1 ± 1.3 | 24.8 ± 2.0 |
| Traits | WT | ny1 | ny2 |
|---|---|---|---|
| Plant height (cm) | 112.0 ± 6.3 a | 102.2 ± 7.5 b | 101.6 ± 6.9 b |
| Days to 50% flowering | 101.0 ± 0.0 b | 107.3 ± 1.5 a | 109.7 ± 1.5 a |
| Flag leaf length (cm) | 35.5 ± 2.6 a | 29.4 ± 4.1 c | 32.1 ± 3.8 b |
| Flag leaf width (cm) | 2.3 ± 0.1 a | 2.1 ± 0.3 b | 2.2 ± 0.2a b |
| Number of panicles | 6.0 ± 0.7 a | 4.0 ± 1.0 b | 4.5 ± 0.9 b |
| Grain length (mm) | 9.4 ± 0.2 a | 9.4 ± 0.7 a | 9.2 ± 0.4 a |
| Grain width (mm) | 3.5 ± 0.1 a | 3.3 ± 0.2 b | 3.4 ± 0.1 ab |
| Grain length to width ratio | 2.7 ± 0.1 b | 2.8 ± 0.3 a | 2.7 ± 0.1 ab |
| Panicle length (cm) | 27.1 ± 1.0 a | 23.6 ± 2.3 b | 23.9 ± 2.1 b |
| Total grains /plant | 768.0 ± 126.1 a | 293.6 ± 118.2 c | 372.8 ± 108.1 b |
| Filled grains/plant | 599.6 ± 94.4 a | 128.4 ± 76.0 c | 186.8 ± 72.3 b |
| 1000-grain weight (g) | 36.3 ± 1.4 a | 26.5 ± 5.5 c | 29.5 ± 2.8 b |
| Grain yield /plant (g) | 21.6 ± 3.4 a | 3.6 ± 2.4 c | 5.5 ± 2.3 b |
| Seed setting (%) | 78.0 ± 4.2 a | 43.3 ± 14.2 b | 49.5 ± 10.2 b |
| Pollen fertility (%) | 93.1 ± 3.3 a | 38.7 ± 36.3 b | 26.7 ± 33.6 b |
| Embryo sac fertility (%) | 90.1 | 85.1 | 87.2 |
| Stage | Wild Type (WT) | ny1 (Mutant Line) | ny2 (Mutant Line) | |||
|---|---|---|---|---|---|---|
| Number of Cells | Abnormal (%) | Number of Cells | Abnormal (%) | Number of Cells | Abnormal (%) | |
| Metaphase I | 688 | 14.7 | 561 | 64.7 | 238 | 66.0 |
| Anaphase I | 139 | 15.8 | 179 | 66.5 | 110 | 62.4 |
| Metaphase II | 149 | 15.4 | 80 | 71.3 | 82 | 90.2 |
| Anaphase II | 37 | 51.4 | 113 | 71.7 | 47 | 87.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamara, N.; Jiao, Y.; Lu, Z.; Aloryi, K.D.; Wu, J.; Liu, X.; Shahid, M.Q. Cytological Observations and Bulked-Segregant Analysis Coupled Global Genome Sequencing Reveal Two Genes Associated with Pollen Fertility in Tetraploid Rice. Int. J. Mol. Sci. 2021, 22, 841. https://doi.org/10.3390/ijms22020841
Kamara N, Jiao Y, Lu Z, Aloryi KD, Wu J, Liu X, Shahid MQ. Cytological Observations and Bulked-Segregant Analysis Coupled Global Genome Sequencing Reveal Two Genes Associated with Pollen Fertility in Tetraploid Rice. International Journal of Molecular Sciences. 2021; 22(2):841. https://doi.org/10.3390/ijms22020841
Chicago/Turabian StyleKamara, Nabieu, Yamin Jiao, Zijun Lu, Kelvin Dodzi Aloryi, Jinwen Wu, Xiangdong Liu, and Muhammad Qasim Shahid. 2021. "Cytological Observations and Bulked-Segregant Analysis Coupled Global Genome Sequencing Reveal Two Genes Associated with Pollen Fertility in Tetraploid Rice" International Journal of Molecular Sciences 22, no. 2: 841. https://doi.org/10.3390/ijms22020841








