Gene Methylation and Silencing of WIF1 Is a Frequent Genetic Abnormality in Mantle Cell Lymphoma
Abstract
:1. Introduction
2. Results
2.1. Expression of WIF1 in MCL Cell Lines and Patient Tumor Samples
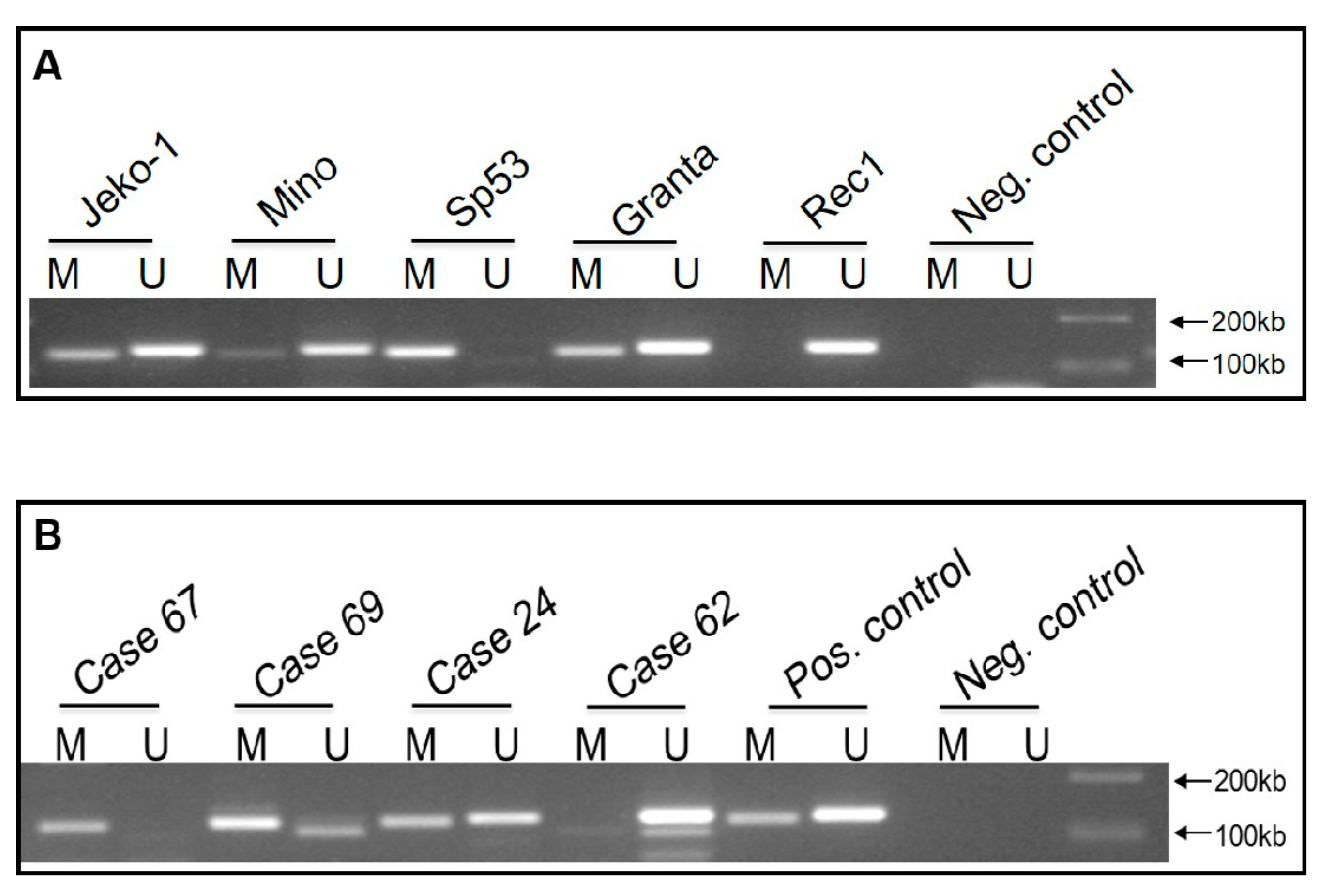
2.2. WIF1 Is Hypermethylated in Most MCL Cell Lines and Tumors
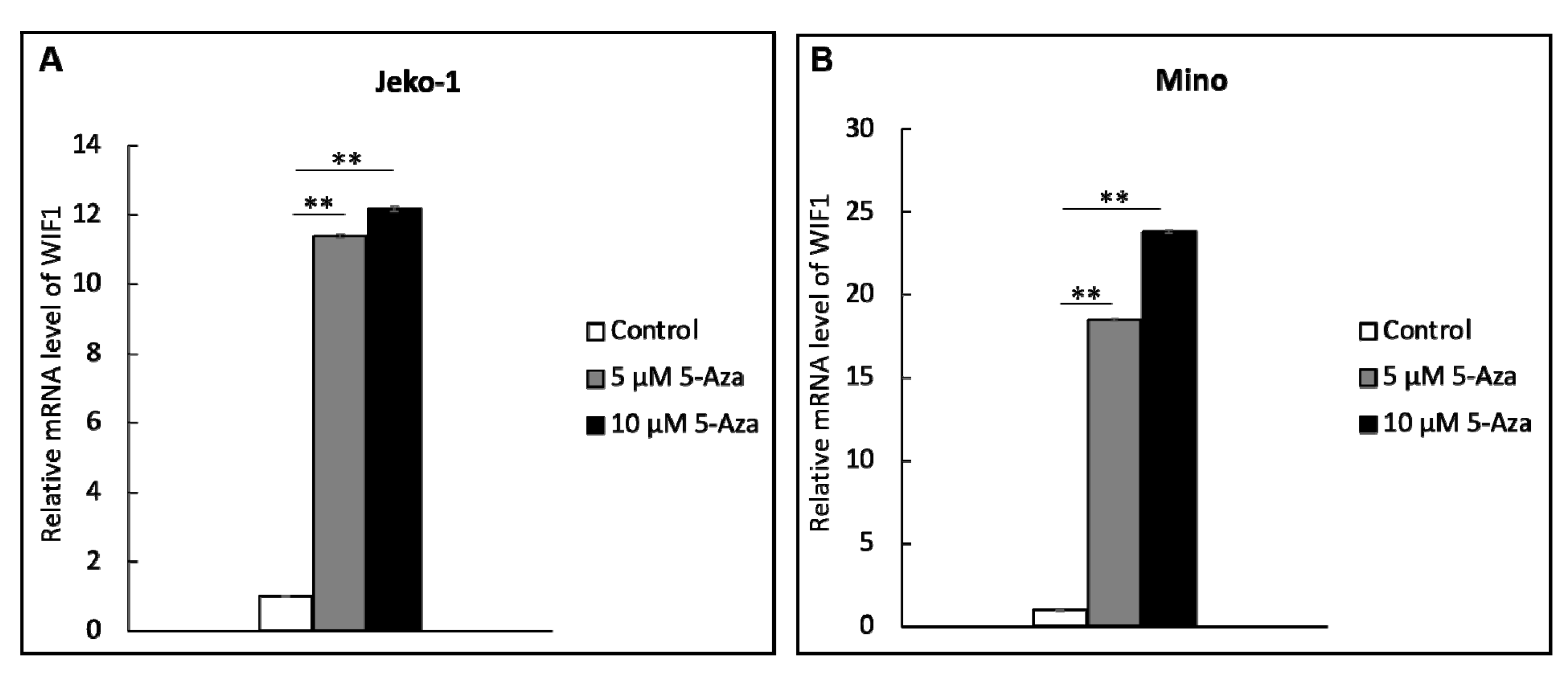
2.3. WIF1 Expression Is Restored by Treatment with Demethylation Agent 5-aza
2.4. WIF1 Decreases β-Catenin and pGSK-3β Expression in MCL Cell Lines
2.5. WIF1 Inhibits MCL Cell Growth and Sensitizes Them to Cytarabine (Ara-C)
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Tumor Samples
4.2. Reverse Transcription-PCR
4.3. Methylation Specific PCR (MSP)
4.4. Treatment with 5-aza-2’-deoxycytidine
4.5. Western Blot Analysis and Antibodies
4.6. Lentiviral Gene Transfection
4.7. MTS Assay and Cell Viability
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parrott, M.; Rule, S.; Kelleher, M.; Wilson, J. A Systematic Review of Treatments of Relapsed/Refractory Mantle Cell Lymphoma. Clin. Lymphoma Myeloma Leuk. 2018, 18, 13–25.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schieber, M.; Gordon, L.I.; Karmali, R. Current overview and treatment of mantle cell lymphoma. Research 2018, 7, 1136. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Galán, P.; Dreyling, M.; Wiestner, A. Mantle cell lymphoma: Biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011, 117, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchakarska, G.; Sola, B. The double dealing of cyclin D1. Cell Cycle 2020, 19, 163–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albero, R.; Enjuanes, A.; Demajo, S.; Castellano, G.; Pinyol, M.; García, N.; Capdevila, C.; Clot, G.; Suárez-Cisneros, H.; Shimada, M.; et al. Cyclin D1 overexpression induces global transcriptional downregulation in lymphoid neoplasms. J. Clin. Investig. 2018, 128, 4132–4147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusté, N.P.; Fernández-Hernández, R.; Cemeli, T.; Mirantes, C.; Pedraza, N.; Rafel, M.; Torres-Rosell, J.; Colomina, N.; Ferrezuelo, F.; Dolcet, X.; et al. Cytoplasmic cyclin D1 regulates cell invasion and metastasis through the phosphorylation of paxillin. Nat. Commun. 2016, 7, 11581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.R.; Joshi, I.; Jin, F.; Al-Saleem, T. Murine model for mantle cell lymphoma. Leukemia 2006, 20, 891–893. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhang, H.F.; Gupta, N.; Alshareef, A.; Wang, Q.; Huang, Y.H.; Lewis, J.T.; Douglas, D.N.; Kneteman, N.M.; Lai, R. A positive feedback loop involving the Wnt/β-catenin/MYC/Sox2 axis defines a highly tumorigenic cell subpopulation in ALK-positive anaplastic large cell lymphoma. J. Hematol. Oncol. 2016, 9, 120. [Google Scholar] [CrossRef] [Green Version]
- Lazarian, G.; Friedrich, C.; Quinquenel, A.; Tran, J.; Ouriemmi, S.; Dondi, E.; Martin, A.; Mihoub, I.; Chiron, D.; Bellanger, C.; et al. Stabilization of beta-catenin upon B-cell receptor signaling promotes NF-kB target genes transcription in mantle cell lymphoma. Oncogene 2020, 39, 2934–2947. [Google Scholar] [CrossRef]
- Van Andel, H.; Kocemba, K.A.; Spaargaren, M.; Pals, S.T. Aberrant Wnt signaling in multiple myeloma: Molecular mechanisms and targeting options. Leukemia 2019, 33, 1063–1075. [Google Scholar] [CrossRef]
- Kotrbová, A.; Ovesná, P.; Gybel’, T.; Radaszkiewicz, T.; Bednaříková, M.; Hausnerová, J.; Jandáková, E.; Minář, L.; Crha, I.; Weinberger, V.; et al. WNT signaling inducing activity in ascites predicts poor outcome in ovarian cancer. Theranostics 2020, 10, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Mathur, R.; Sehgal, L.; Braun, F.K.; Berkova, Z.; Romaguerra, J.; Wang, M.; Rodriguez, M.A.; Fayad, L.; Neelapu, S.S.; Samaniego, F.; et al. Targeting Wnt pathway in mantle cell lymphoma-initiating cells. J. Hematol. Oncol. 2015, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, E.A.; Hooker, C.M.; McDevitt, M.A.; Karp, J.E.; Herman, J.G.; Carraway, H.E. Acute Myeloid Leukemia Is Characterized by Wnt Pathway Inhibitor Promoter Methylation. Blood 2008, 112, 2253. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, H.; Zhang, G.; Xue, X. Targeting the canonical Wnt/β-catenin pathway in cancer radioresistance: Updates on the molecular mechanisms. J. Cancer Res. Ther. 2019, 15, 272–277. [Google Scholar] [PubMed]
- Lecarpentier, Y.; Schussler, O.; Hébert, J.L.; Vallée, A. Multiple Targets of the Canonical WNT/β-Catenin Signaling in Cancers. Front. Oncol. 2019, 9, 1248. [Google Scholar] [CrossRef]
- Chung, R.; Peters, A.C.; Armanious, H.; Anand, M.; Gelebart, P.; Lai, R. Biological and clinical significance of GSK-3beta in mantle cell lymphoma--an immunohistochemical study. Int. J. Clin. Exp. Pathol. 2010, 3, 244–253. [Google Scholar]
- Gelebart, P.; Anand, M.; Armanious, H.; Peters, A.C.; Bard, J.D.; Amin, H.M.; Lai, R. Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood 2008, 112, 5171–5179. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, J.; Shen, Y.; Liu, Y.; Fu, K.; Jaffe, E.S.; Liu, C.; Liu, Z.; Lachel, C.M.; Deffenbacher, K.; Greiner, T.C.; et al. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood 2012, 119, 4939–4948. [Google Scholar] [CrossRef]
- Morgan, A.E.; Davies, T.J.; Mc Auley, M.T. The role of DNA methylation in ageing and cancer. Proc. Nutr. Soc. 2018, 77, 412–422. [Google Scholar] [CrossRef]
- Bouras, E.; Karakioulaki, M.; Bougioukas, K.I.; Aivaliotis, M.; Tzimagiorgis, G.; Chourdakis, M. Gene promoter methylation and cancer: An umbrella review. Gene 2019, 710, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.Y.; Chen, C.X.; Li, L. Hypermethylation of tumor suppressor genes is a risk factor for poor prognosis in ovarian cancer: A meta-analysis. Medicine 2019, 98, e14588. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Issa, J.J.; Kropf, P. DNA Hypomethylating Drugs in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026948. [Google Scholar] [CrossRef] [Green Version]
- Dombret, H.; Itzykson, R. How and when to decide between epigenetic therapy and chemotherapy in patients with AML. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Bohl, S.R.; Bullinger, L.; Rücker, F.G. Epigenetic therapy: Azacytidine and decitabine in acute myeloid leukemia. Expert Rev. Hematol. 2018, 11, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Poggi, L.; Casarosa, S.; Carl, M. An Eye on the Wnt Inhibitory Factor Wif1. Front. Cell Dev. Biol. 2018, 6, 167. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Wu, J.; Cai, Y. Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 438, 673–679. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Z.; Ding, Y.; Zhang, P.; Wang, J.; Zhang, J.; Wang, H. Promoter methylation of WNT inhibitory factor-1 may be associated with the pathogenesis of multiple human tumors. J. Cancer Res. Ther. 2018, 14, 381–387. [Google Scholar]
- Tang, Q.; Zhao, H.; Yang, B.; Li, L.; Shi, Q.; Jiang, C.; Liu, H. WIF-1 gene inhibition and Wnt signal transduction pathway activation in NSCLC tumorigenesis. Oncol. Lett. 2017, 13, 1183–1188. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Fu, J.; Bi, H.; Ge, A.; Xia, T.; Liu, Y.; Sun, H.; Li, D.; Zhao, Y. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer 2019, 19, 1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Hou, C.; Wang, H.; Liang, T.; Zhu, L. Hypermethylation of WIF1 and its inhibitory role in the tumor growth of endometrial adenocarcinoma. Mol. Med. Rep. 2017, 16, 7497–7503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, J.H.; Sloan, S.; Scherle, P.; Vaddi, K.; Sif, S.; Lapalombella, R.; A Baiocchi, R. PRMT5 Is a Key Epigenetic Regulator That Promotes Transcriptional Activation in Mantle Cell Lymphoma By Regulating the Lysine Methyltransferase SETD7 and MLL1 Activity. Blood 2019, 134 (Suppl. 1), 2777. [Google Scholar] [CrossRef]
- Lyu, X.; Li, J.; Yun, X.; Huang, R.; Deng, X.; Wang, Y.; Chen, Y.; Xiao, G. miR-181a-5p, an inducer of Wnt-signaling, facilitates cell proliferation in acute lymphoblastic leukemia. Oncol. Rep. 2017, 37, 1469–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, V.; Agirre, X.; Jiménez-Velasco, A.; José-Enériz, E.S.; Cordeu, L.; Garate, L.; Vilas-Zornoza, A.; Castillejo, J.A.; Heiniger, A.; Prosper, F.; et al. Methylation status of Wnt signaling pathway genes affects the clinical outcome of Philadelphia-positive acute lymphoblastic leukemia. Cancer Sci. 2008, 99, 1865–1868. [Google Scholar] [CrossRef]
- Chim, C.S.; Fung, T.K.; Wong, K.F.; Lau, J.S.; Liang, R. Infrequent Wnt inhibitory factor-1 (Wif-1) methylation in chronic lymphocytic leukemia. Leuk. Res. 2006, 30, 1135–1139. [Google Scholar] [CrossRef]
- Taniguchi, H.; Yamamoto, H.; Hirata, T.; Miyamoto, N.; Oki, M.; Nosho, K.; Adachi, Y.; Endo, T.; Imai, K.; Shinomura, Y. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene 2005, 24, 7946–7952. [Google Scholar] [CrossRef] [Green Version]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [Green Version]
- Mazieres, J.; He, B.; You, L.; Xu, Z.; Lee, A.Y.; Mikami, I.; Reguart, N.; Rosell, R.; McCormick, F.; Jablons, D.M. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004, 64, 4717–4720. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Park, J.Y.; Kim, D.S. Wif1 hypermethylation as unfavorable prognosis of non-small cell lung cancers with EGFR mutation. Mol. Cells 2013, 36, 69–73. [Google Scholar] [CrossRef] [Green Version]



| Methylation Status | Low pGSK-3β | High pGSK-3β | p Value |
|---|---|---|---|
| WIF1 methylation detected | 6 | 15 | 0.038 |
| WIF1 methylation NOT detected. | 6 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshareef, A.; Peters, A.C.; Gélébart, P.; Chen, W.; Lai, R. Gene Methylation and Silencing of WIF1 Is a Frequent Genetic Abnormality in Mantle Cell Lymphoma. Int. J. Mol. Sci. 2021, 22, 893. https://doi.org/10.3390/ijms22020893
Alshareef A, Peters AC, Gélébart P, Chen W, Lai R. Gene Methylation and Silencing of WIF1 Is a Frequent Genetic Abnormality in Mantle Cell Lymphoma. International Journal of Molecular Sciences. 2021; 22(2):893. https://doi.org/10.3390/ijms22020893
Chicago/Turabian StyleAlshareef, Abdulraheem, Anthea C. Peters, Pascal Gélébart, Will Chen, and Raymond Lai. 2021. "Gene Methylation and Silencing of WIF1 Is a Frequent Genetic Abnormality in Mantle Cell Lymphoma" International Journal of Molecular Sciences 22, no. 2: 893. https://doi.org/10.3390/ijms22020893
APA StyleAlshareef, A., Peters, A. C., Gélébart, P., Chen, W., & Lai, R. (2021). Gene Methylation and Silencing of WIF1 Is a Frequent Genetic Abnormality in Mantle Cell Lymphoma. International Journal of Molecular Sciences, 22(2), 893. https://doi.org/10.3390/ijms22020893






